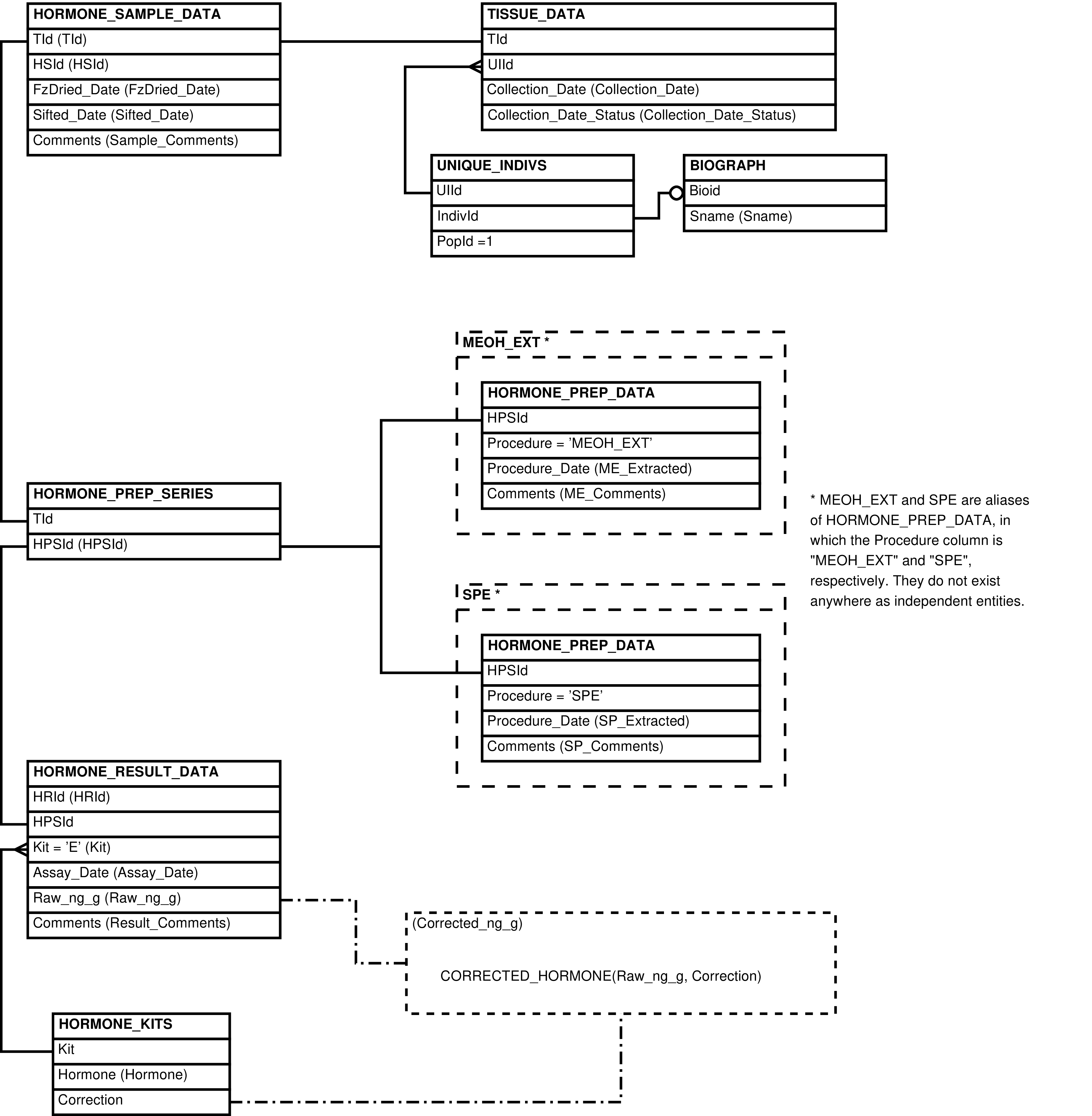
Contains one row for every sample whose estrogen
concentration has been measured by any kit with a known
correction factor. Results from kits whose HORMONE_KITS.Correction is NULL are
omitted.
Tip
Use this view to see estrogen concentration in all the hormone samples. It joins all the pertinent tables together to gather information, and omits results that are not considered reliable.
This view includes a "Hormone" column that indicates
which hormone was measured in each assay. By definition, this
column will be E in every row,
so it may seem odd to include the row at all. The column is
retained as a courtesy to users, especially for those who
might unify the rows from this view with rows of other,
similar views (e.g. GLUCOCORTICOIDS, PROGESTERONES, etc.).
Figure 6.24. Query Defining the ESTROGENS View
SELECT hormone_sample_data.tid
, hormone_prep_series.hpsid
, hormone_result_data.hrid
, hormone_sample_data.hsid
, biograph.sname
, tissue_data.collection_date
, tissue_data.collection_date_status AS collection_date_status
, hormone_sample_data.fzdried_date AS fzdried_date
, hormone_sample_data.sifted_date AS sifted_date
, meoh_ext.procedure_date AS me_extracted
, spe.procedure_date AS sp_extracted
, hormone_result_data.raw_ng_g AS raw_ng_g
, corrected_hormone(hormone_result_data.raw_ng_g, hormone_kits.correction) AS corrected_ng_g
, hormone_result_data.assay_date
, hormone_kits.hormone AS hormone
, hormone_result_data.kit AS kit
, hormone_sample_data.comments AS sample_comments
, hormone_result_data.comments AS result_comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ
JOIN hormone_prep_series
ON hormone_prep_series.tid = hormone_sample_data.tid
JOIN hormone_result_data
ON hormone_result_data.hpsid = hormone_prep_series.hpsid
JOIN hormone_kits
ON hormone_kits.kit = hormone_result_data.kit
AND hormone_kits.correction IS NOT NULL
AND hormone_kits.hormone = 'E'
LEFT JOIN hormone_prep_data AS meoh_ext
ON meoh_ext.procedure = 'MEOH_EXT'
AND meoh_ext.hpsid = hormone_prep_series.hpsid
LEFT JOIN hormone_prep_data AS spe
ON spe.procedure = 'SPE'
AND spe.hpsid = hormone_prep_series.hpsid;
Table 6.11. Columns in the ESTROGENS View
| Column | From | Description |
|---|---|---|
| TId | HORMONE_SAMPLE_DATA.TId | Identifier of the HORMONE_SAMPLE_DATA row and tissue sample |
| HPSId | HORMONE_PREP_SERIES.HPSId | Identifier of the prep series |
| HRId | HORMONE_RESULT_DATA.HRId | Identifier of the assay that generated this result |
| HSId | HORMONE_SAMPLE_DATA.HSId | User-generated identifier for the tissue sample |
| Sname | BIOGRAPH.Sname | Sname of the individual from whom this sample came |
| Collection_Date | TISSUE_DATA.Collection_Date | Date the tissue sample was collected |
| Collection_Date_Status | TISSUE_DATA.Collection_Date_Status | The status of this Collection_Date |
| FzDried_Date | HORMONE_SAMPLE_DATA.FzDried_Date | Date the sample was freeze-dried |
| Sifted_Date | HORMONE_SAMPLE_DATA.Sifted_Date | Date the freeze-dried sample was sifted |
| ME_Extracted | HORMONE_PREP_DATA.Procedure_Date | Date of methanol extraction, in prep for this result |
| SP_Extracted | HORMONE_PREP_DATA.Procedure_Date | Date of solid-phase extraction, in prep for this result |
| Raw_ng_g | HORMONE_RESULT_DATA.Raw_ng_g | The "raw" concentration determined in this assay |
| Corrected_ng_g | CORRECTED_HORMONE(HORMONE_RESULT_DATA.Raw_ng_g, HORMONE_KITS.Correction) | The corrected concentration, according to the related HORMONE_KITS.Correction |
| Assay_Date | HORMONE_RESULT_DATA.Assay_Date | Date of this assay |
| Hormone | HORMONE_KITS.Hormone | Code for the hormone whose concentration was assayed |
| Kit | HORMONE_RESULT_DATA.Kit | Code for the kit used in this assay |
| Sample_Comments | HORMONE_SAMPLE_DATA.Comments | Comments about the hormone sample |
| ME_Comments | HORMONE_PREP_DATA.Comments | Comments about the methanol extraction |
| SP_Comments | HORMONE_PREP_DATA.Comments | Comments about the solid-phase extraction |
| Result_Comments | HORMONE_RESULT_DATA.Comments | Comments about the assay |
Contains one row for every sample whose glucocorticoid
concentration has been measured by any kit with a known
correction factor. Results from kits whose HORMONE_KITS.Correction is NULL are
omitted.
Tip
Use this view to see glucocorticoid concentration in all the hormone samples. It joins all the pertinent tables together to gather information, and omits results that are not considered reliable.
This view includes a "Hormone" column that indicates
which hormone was measured in each assay. By definition, this
column will be GC in every
row, so it may seem odd to include the row at all. The column
is retained as a courtesy to users, especially for those who
might unify the rows from this view with rows of other,
similar views (e.g. ESTROGENS, PROGESTERONES, etc.).
Figure 6.26. Query Defining the GLUCOCORTICOIDS View
SELECT hormone_sample_data.tid
, hormone_prep_series.hpsid
, hormone_result_data.hrid
, hormone_sample_data.hsid
, biograph.sname
, tissue_data.collection_date
, tissue_data.collection_date_status AS collection_date_status
, hormone_sample_data.fzdried_date AS fzdried_date
, hormone_sample_data.sifted_date AS sifted_date
, meoh_ext.procedure_date AS me_extracted
, spe.procedure_date AS sp_extracted
, hormone_result_data.raw_ng_g AS raw_ng_g
, corrected_hormone(hormone_result_data.raw_ng_g, hormone_kits.correction) AS corrected_ng_g
, hormone_result_data.assay_date
, hormone_kits.hormone AS hormone
, hormone_result_data.kit AS kit
, hormone_sample_data.comments AS sample_comments
, hormone_result_data.comments AS result_comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ
JOIN hormone_prep_series
ON hormone_prep_series.tid = hormone_sample_data.tid
JOIN hormone_result_data
ON hormone_result_data.hpsid = hormone_prep_series.hpsid
JOIN hormone_kits
ON hormone_kits.kit = hormone_result_data.kit
AND hormone_kits.correction IS NOT NULL
AND hormone_kits.hormone = 'GC'
LEFT JOIN hormone_prep_data AS meoh_ext
ON meoh_ext.procedure = 'MEOH_EXT'
AND meoh_ext.hpsid = hormone_prep_series.hpsid
LEFT JOIN hormone_prep_data AS spe
ON spe.procedure = 'SPE'
AND spe.hpsid = hormone_prep_series.hpsid;
Table 6.12. Columns in the GLUCOCORTICOIDS View
| Column | From | Description |
|---|---|---|
| TId | HORMONE_SAMPLE_DATA.TId | Identifier of the HORMONE_SAMPLE_DATA row and tissue sample |
| HPSId | HORMONE_PREP_SERIES.HPSId | Identifier of the prep series |
| HRId | HORMONE_RESULT_DATA.HRId | Identifier of the assay that generated this result |
| HSId | HORMONE_SAMPLE_DATA.HSId | User-generated identifier for the tissue sample |
| Sname | BIOGRAPH.Sname | Sname of the individual from whom this sample came |
| Collection_Date | TISSUE_DATA.Collection_Date | Date the tissue sample was collected |
| Collection_Date_Status | TISSUE_DATA.Collection_Date_Status | The status of this Collection_Date |
| FzDried_Date | HORMONE_SAMPLE_DATA.FzDried_Date | Date the sample was freeze-dried |
| Sifted_Date | HORMONE_SAMPLE_DATA.Sifted_Date | Date the freeze-dried sample was sifted |
| ME_Extracted | HORMONE_PREP_DATA.Procedure_Date | Date of methanol extraction, in prep for this result |
| SP_Extracted | HORMONE_PREP_DATA.Procedure_Date | Date of solid-phase extraction, in prep for this result |
| Raw_ng_g | HORMONE_RESULT_DATA.Raw_ng_g | The "raw" concentration determined in this assay |
| Corrected_ng_g | CORRECTED_HORMONE(HORMONE_RESULT_DATA.Raw_ng_g, HORMONE_KITS.Correction) | The corrected concentration, according to the related HORMONE_KITS.Correction |
| Assay_Date | HORMONE_RESULT_DATA.Assay_Date | Date of this assay |
| Hormone | HORMONE_KITS.Hormone | Code for the hormone whose concentration was assayed |
| Kit | HORMONE_RESULT_DATA.Kit | Code for the kit used in this assay |
| Sample_Comments | HORMONE_SAMPLE_DATA.Comments | Comments about the hormone sample |
| ME_Comments | HORMONE_PREP_DATA.Comments | Comments about the methanol extraction |
| SP_Comments | HORMONE_PREP_DATA.Comments | Comments about the solid-phase extraction |
| Result_Comments | HORMONE_RESULT_DATA.Comments | Comments about the assay |
Contains one row for every laboratory preparation that was performed on a sample. This view includes columns from BIOGRAPH, HORMONE_PREP_DATA, HORMONE_PREP_SERIES, HORMONE_SAMPLE_DATA, TISSUE_DATA, and UNIQUE_INDIVS in order to portray information in a more user-friendly format. This view is also useful for uploading new data.
Tip
Use this view instead of the HORMONE_PREP_DATA table.
Figure 6.28. Query Defining the HORMONE_PREPS View
SELECT hormone_sample_data.tid AS tid
, hormone_sample_data.hsid AS hsid
, unique_indivs.individ AS individ
, biograph.sname AS sname
, hormone_prep_series.hpsid AS hpsid
, hormone_prep_series.series AS series
, hormone_prep_data.hpid AS hpid
, hormone_prep_data.procedure AS procedure
, hormone_prep_data.procedure_date AS procedure_date
, hormone_prep_data.comments AS comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ
JOIN hormone_prep_series
ON hormone_prep_series.tid = hormone_sample_data.tid
JOIN hormone_prep_data
ON hormone_prep_data.hpsid = hormone_prep_series.hpsid;
Table 6.13. Columns in the HORMONE_PREPS View
| Column | From | Description |
|---|---|---|
| TId | HORMONE_SAMPLE_DATA.TId | Identifier of the HORMONE_SAMPLE_DATA row and tissue sample |
| HSId | HORMONE_SAMPLE_DATA.HSId | User-generated identifier for the tissue sample |
| IndivId | UNIQUE_INDIVS.IndivId | Name/ID for the source individual |
| Sname | BIOGRAPH.Sname | Sname of the source individual |
| HPSId | HORMONE_PREP_SERIES.HPSId | Identifier of the series to which this prep belongs |
| Series | HORMONE_PREP_SERIES.Series | Identifier for the prep series for this sample |
| HPId | HORMONE_PREP_DATA.HPId | Identifier for the HORMONE_PREP_DATA row |
| Procedure | HORMONE_PREP_DATA.Procedure | Code indicating what was done |
| Procedure_Date | HORMONE_PREP_DATA.Procedure_Date | Date that this prep was performed |
| Comments | HORMONE_PREP_DATA.Comments | Miscellaneous notes/comments about this prep |
Inserting a row into HORMONE_PREPS inserts a row in HORMONE_PREP_DATA, as expected. A new HORMONE_PREP_SERIES row may also be inserted, as described below.
When identifying the related tissue sample, either or both of the TId and HSId columns must be provided. If both, they must be related in HORMONE_SAMPLE_DATA.
It is not necessary to provide IndivId or Sname values. Any such values that are provided must match the related values for the provided TId and/or HSId.
If HPSId is provided, it must already be an HPSId value in HORMONE_PREP_SERIES, and its related TId must match the provided TId or be related to the provided HSId.
If a row’s series has not yet been added to HORMONE_PREP_SERIES, this view can add it. When no HPSId is provided, the view will use the provided Series and either TId or HSId values to determine if there is already such a row in HORMONE_PREP_SERIES. If no such HORMONE_PREP_SERIES row is found, then those values are used to create a new HORMONE_PREP_SERIES row. The inserted HORMONE_PREP_DATA.HPSId is either that of the found row or of the newly-created one.
Updating a row in HORMONE_PREPS updates the underlying row in HORMONE_PREP_DATA, as expected.
If both TId and HSId are updated, they must be related in HORMONE_SAMPLE_DATA. If either or both of those columns are updated, either or both of the Series and HPSId columns must also be updated.
To update the HORMONE_PREP_DATA.HPSId column, the HPSId can be updated directly, or the Series can be updated alone. If the Series is updated without the HPSId, this view will use the Series and the TId to look up the correct HPSId from HORMONE_PREP_SERIES. If changing the HPSId also requires a change to the TId and HSId, then an appropriate update to either or both of those columns must be provided at the same time as the update to the HPSId.
If any of the TId, HSId, HPSId, or Series columns
are changed, there must already be a HORMONE_PREP_SERIES row containing the new
values. Unlike on INSERT, this view
will not create a new series in HORMONE_PREP_SERIES on
UPDATE.
Attempts to update the IndivId or Sname columns will return an error.
Tip
To change either of these values, you should update only the TId or HSId column, or update the related TISSUE_DATA row.
Deleting a row from HORMONE_PREPS deletes the underlying row in HORMONE_PREP_DATA, as expected. The related row in HORMONE_PREP_SERIES is unaffected.
Contains one row for every hormone assay result for a sample. That is, every HORMONE_RESULT_DATA row. This view includes columns from BIOGRAPH, HORMONE_PREP_SERIES, HORMONE_RESULT_DATA, HORMONE_SAMPLE_DATA, TISSUE_DATA, and UNIQUE_INDIVS in order to portray information in a more user-friendly format. This view is also useful for uploading new data.
Tip
Use this view instead of the HORMONE_RESULT_DATA table.
Figure 6.30. Query Defining the HORMONE_RESULTS View
SELECT hormone_sample_data.tid AS tid
, hormone_sample_data.hsid AS hsid
, unique_indivs.individ AS individ
, biograph.sname AS sname
, hormone_prep_series.hpsid AS hpsid
, hormone_prep_series.series AS series
, hormone_result_data.hrid AS hrid
, hormone_kits.hormone AS hormone
, hormone_result_data.kit AS kit
, hormone_result_data.assay_date AS assay_date
, hormone_result_data.grams_used AS grams_used
, hormone_result_data.raw_ng_g AS raw_ng_g
, corrected_hormone(hormone_result_data.raw_ng_g, hormone_kits.correction) AS corrected_ng_g
, hormone_result_data.comments AS comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ
JOIN hormone_prep_series
ON hormone_prep_series.tid = hormone_sample_data.tid
JOIN hormone_result_data
ON hormone_result_data.hpsid = hormone_prep_series.hpsid
JOIN hormone_kits
ON hormone_kits.kit = hormone_result_data.kit;
Table 6.14. Columns in the HORMONE_RESULTS View
| Column | From | Description |
|---|---|---|
| TId | HORMONE_SAMPLE_DATA.TId | Identifier of the HORMONE_SAMPLE_DATA row and tissue sample |
| HSId | HORMONE_SAMPLE_DATA.HSId | User-generated identifier for the tissue sample |
| IndivId | UNIQUE_INDIVS.IndivId | Name/ID for the source individual |
| Sname | BIOGRAPH.Sname | Sname of the source individual |
| HPSId | HORMONE_PREP_SERIES.HPSId | Identifier of the series to which this prep belongs |
| Series | HORMONE_PREP_SERIES.Series | Identifier for the prep series for this sample |
| HRId | HORMONE_RESULT_DATA.HRId | Identifier of the HORMONE_RESULT_DATA row |
| Hormone | HORMONE_KITS.Hormone | The hormone that was measured in this result |
| Kit | HORMONE_RESULT_DATA.Kit | The kit used to perform this assay |
| Assay_Date | HORMONE_RESULT_DATA.Assay_Date | The date of this assay |
| Grams_Used | HORMONE_RESULT_DATA.Grams_Used | The mass of tissue in grams that was consumed to generate this result |
| Raw_ng_g | HORMONE_RESULT_DATA.Raw_ng_g | The "raw" (uncorrected) concentration of this hormone in ng/g according to this assay |
| Corrected_ng_g | CORRECTED_HORMONE(HORMONE_RESULT_DATA.Raw_ng_g, HORMONE_KITS.Correction) | The corrected concentration, according to the related HORMONE_KITS.Correction |
| Comments | HORMONE_RESULT_DATA.Comments | Miscellaneous notes/comments about this result |
Inserting a row into HORMONE_RESULTS inserts a row in HORMONE_RESULT_DATA, as expected.
When identifying the related tissue sample, either the TId or HSId must be provided. If both are provided, they must be related in HORMONE_SAMPLE_DATA.
It is not necessary to provide IndivId or Sname values. Any such values that are provided must match the related values for the provided TId or HSId.
When identifying the HORMONE_RESULT_DATA.HPSId to insert, either or both of the HPSId and Series columns must be provided[249].
Any provided HSId, TId, HPSId, and/or Series values must be related in HORMONE_SAMPLE_DATA and HORMONE_PREP_SERIES.
It is not necessary to provide a Hormone value. If one is provided, it must match the related HORMONE_KITS.Hormone for the provided Kit.
It is not necessary to provide a Corrected_ng_g value, as this is a calculated column. If one is provided, it must match the value that is calculated by the corrected_hormone() function with the provided Raw_ng_g and the related HORMONE_KITS.Correction.
Updating a row in HORMONE_RESULTS updates the underlying row in HORMONE_RESULT_DATA, as expected.
If both TId and HSId are updated, they must be related in HORMONE_SAMPLE_DATA. If either or both of those columns are updated, either or both of the Series and HPSId columns must also be updated.
To update the HORMONE_RESULT_DATA.HPSId column, the HPSId can be updated directly, or the Series can be updated alone. If the Series is updated without the HPSId, this view will use the Series and the TId to look up the correct HPSId from HORMONE_PREP_SERIES. If changing the HPSId also requires a change to the TId and HSId, then an appropriate update to either or both of those columns must be provided at the same time as the update to the HPSId.
If any of the TId, HSId, HPSId, or Series columns are changed, there must already be a HORMONE_PREP_SERIES row containing the new values.
Attempts to update the IndivId or Sname columns will return an error.
Tip
To change either of these values, you should update only the TId or HSId column, or update the related TISSUE_DATA row.
Attempts to update the Hormone column will return an error.
Tip
To change this value, you should update the Kit column.
If the Corrected_ng_g is updated, the new value must match the value that is calculated by the corrected_hormone() function with the row's Raw_ng_g and the related HORMONE_KITS.Correction, which will only happen if either or both of the Raw_ng_g and Kit columns is also updated.
Tip
To change the concentration for a row, update the Raw_ng_g and let the system determine the corrected concentration.
Deleting a row from HORMONE_RESULTS deletes the underlying row from HORMONE_RESULT_DATA, as expected.
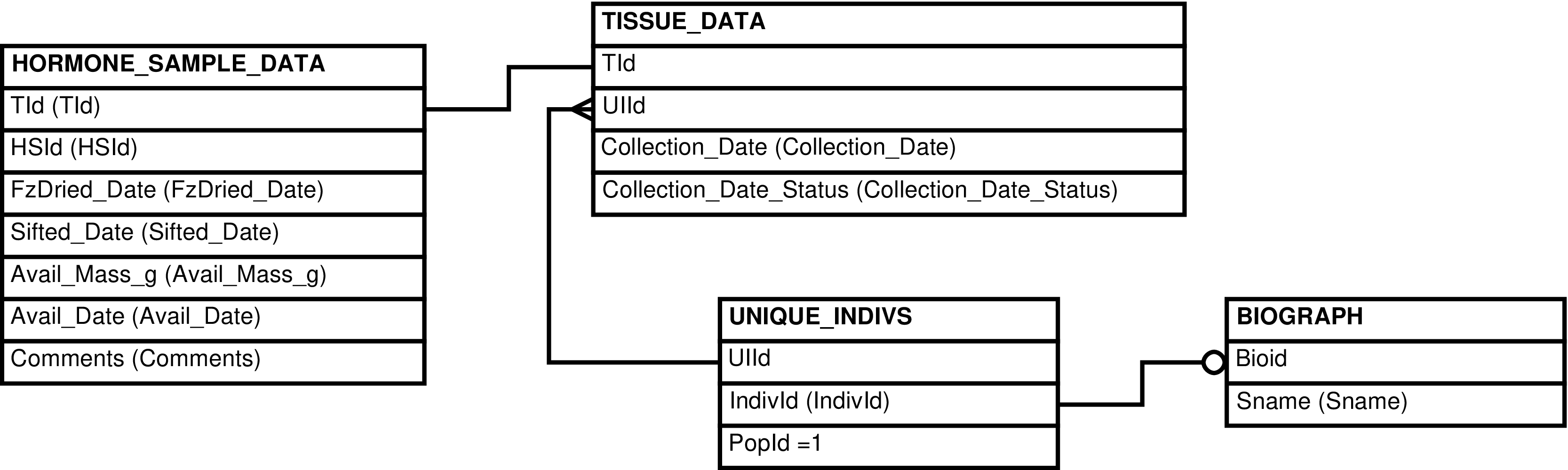
Contains one row for every tissue sample that has undergone any hormone analysis. That is, for every HORMONE_SAMPLE_DATA row. This view includes columns from BIOGRAPH, HORMONE_SAMPLE_DATA, TISSUE_DATA, and UNIQUE_INDIVS in order to portray information in a more user-friendly format. This view is also useful for uploading new data.
Tip
Use this view instead of the HORMONE_SAMPLE_DATA table.
Figure 6.32. Query Defining the HORMONE_SAMPLES View
SELECT hormone_sample_data.tid AS tid
, hormone_sample_data.hsid AS hsid
, unique_indivs.individ AS individ
, biograph.sname AS sname
, tissue_data.collection_date AS collection_date
, tissue_data.collection_date_status AS collection_date_status
, hormone_sample_data.fzdried_date AS fzdried_date
, hormone_sample_data.sifted_date AS sifted_date
, hormone_sample_data.avail_mass_g AS avail_mass_g
, hormone_sample_data.avail_date AS avail_date
, hormone_sample_data.comments AS comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ;
Table 6.15. Columns in the HORMONE_SAMPLES View
| Column | From | Description |
|---|---|---|
| TId | HORMONE_SAMPLE_DATA.TId | Identifier of the HORMONE_SAMPLE_DATA row and tissue sample |
| HSId | HORMONE_SAMPLE_DATA.HSId | User-generated identifier for the tissue sample |
| IndivId | UNIQUE_INDIVS.IndivId | Name/ID for the source individual |
| Sname | BIOGRAPH.Sname | Sname of the source individual |
| Collection_Date | TISSUE_DATA.Collection_Date | Date the tissue sample was collected |
| Collection_Date_Status | TISSUE_DATA.Collection_Date_Status | The status of this Collection_Date |
| FzDried_Date | HORMONE_SAMPLE_DATA.FzDried_Date | Date the sample was freeze-dried |
| Sifted_Date | HORMONE_SAMPLE_DATA.Sifted_Date | Date the freeze-dried sample was sifted |
| Avail_Mass_g | HORMONE_SAMPLE_DATA.Avail_Mass_g | Amount of sample (in g) remaining in the tube, as of the Avail_Date |
| Avail_Date | HORMONE_SAMPLE_DATA.Avail_Date | Date that the Avail_Mass_g was determined |
| Comments | HORMONE_SAMPLE_DATA.Comments | Miscellaneous notes/comments about this sample |
Inserting a row into HORMONE_SAMPLES inserts a row in HORMONE_SAMPLE_DATA, as expected.
It is not necessary to provide IndivId, Sname, or Collection_Date values. Any such values that are provided must match the related values for the provided TId.
Updating a row in HORMONE_SAMPLES updates the underlying row in HORMONE_SAMPLE_DATA, as expected.
Attempts to update the IndivId, Sname, or Collection_Date columns will return an error.
Tip
To change any of these values for a sample, update the related TISSUE_DATA row.
Deleting a row from HORMONE_SAMPLES deletes the underlying row in HORMONE_SAMPLE_DATA, as expected. The related row in TISSUE_DATA is unaffected.
This view shows all the hybrid morphology scores, one line per report.
Figure 6.34. Query Defining the HYBRIDMORPH_SCORES View
WITH concat_observers AS (SELECT hmrid
, string_agg(observer, '/' ORDER BY hmoid) AS observers
FROM hybridmorph_observers
GROUP BY hmrid)
SELECT hybridmorph_reports.hmrid AS hmrid
, hybridmorph_reports.sname AS sname
, hybridmorph_reports.date AS date
, hybridmorph_reports.grp AS grp
, concat_observers.observers AS observers
, color.score AS color
, hair_leng.score AS hair_leng
, body_shape.score AS body_shape
, head_shape.score AS head_shape
, tail_leng.score AS tail_leng
, tail_bend.score AS tail_bend
, muzzle.score AS muzzle
FROM hybridmorph_reports
LEFT JOIN concat_observers
ON concat_observers.hmrid = hybridmorph_reports.hmrid
LEFT JOIN hybridmorph_score_data AS color
ON color.hmrid = hybridmorph_reports.hmrid
AND color.trait = 'COLOR'
LEFT JOIN hybridmorph_score_data AS hair_leng
ON hair_leng.hmrid = hybridmorph_reports.hmrid
AND hair_leng.trait = 'HAIR_LENG'
LEFT JOIN hybridmorph_score_data AS body_shape
ON body_shape.hmrid = hybridmorph_reports.hmrid
AND body_shape.trait = 'BODY_SHAPE'
LEFT JOIN hybridmorph_score_data AS head_shape
ON head_shape.hmrid = hybridmorph_reports.hmrid
AND head_shape.trait = 'HEAD_SHAPE'
LEFT JOIN hybridmorph_score_data AS tail_leng
ON tail_leng.hmrid = hybridmorph_reports.hmrid
AND tail_leng.trait = 'TAIL_LENG'
LEFT JOIN hybridmorph_score_data AS tail_bend
ON tail_bend.hmrid = hybridmorph_reports.hmrid
AND tail_bend.trait = 'TAIL_BEND'
LEFT JOIN hybridmorph_score_data AS muzzle
ON muzzle.hmrid = hybridmorph_reports.hmrid
AND muzzle.trait = 'MUZZLE';
Table 6.16. Columns in the HYBRIDMORPH_SCORES View
| Column | From | Description |
|---|---|---|
| HMRId | HYBRIDMORPH_REPORTS.HMRId | Identifier for the report. |
| Sname | HYBRIDMORPH_REPORTS.Sname | The scored individual. |
| Date | HYBRIDMORPH_REPORTS.Date | The date of the scoring. |
| Grp | HYBRIDMORPH_REPORTS.Grp | The individual's group on this date, according to the observer(s). |
| Observers | HYBRIDMORPH_OBSERVERS.Observer | All of this report's observers, concatenated
together and separated by a
"/". If no
related observers, then NULL. |
| Color | HYBRIDMORPH_SCORE_DATA.Score | The individual's "Coat Color" score. |
| Hair_Leng | HYBRIDMORPH_SCORE_DATA.Score | The individual's "Hair Length" score. |
| Body_Shape | HYBRIDMORPH_SCORE_DATA.Score | The individual's "Body Shape" score. |
| Head_Shape | HYBRIDMORPH_SCORE_DATA.Score | The individual's "Head Shape" score. |
| Tail_Leng | HYBRIDMORPH_SCORE_DATA.Score | The individual's "Tail Length" score. |
| Tail_Bend | HYBRIDMORPH_SCORE_DATA.Score | The individual's "Tail_Bend" score. |
| Muzzle | HYBRIDMORPH_SCORE_DATA.Score | The individual's "Muzzle" score. |
Inserting a row into this view inserts rows into
HYBRIDMORPH_REPORTS, HYBRIDMORPH_OBSERVERS, and HYBRIDMORPH_SCORE_DATA as expected. If one of
the "score" columns is NULL, a HYBRIDMORPH_SCORE_DATA row with that column's
Trait is NOT added.
Updating a row in this view updates rows in HYBRIDMORPH_REPORTS, HYBRIDMORPH_OBSERVERS, and HYBRIDMORPH_SCORE_DATA as expected.
The HMRId cannot be changed.
Deleting a row from this view deletes from HYBRIDMORPH_REPORTS, HYBRIDMORPH_OBSERVERS, and HYBRIDMORPH_SCORE_DATA as expected.
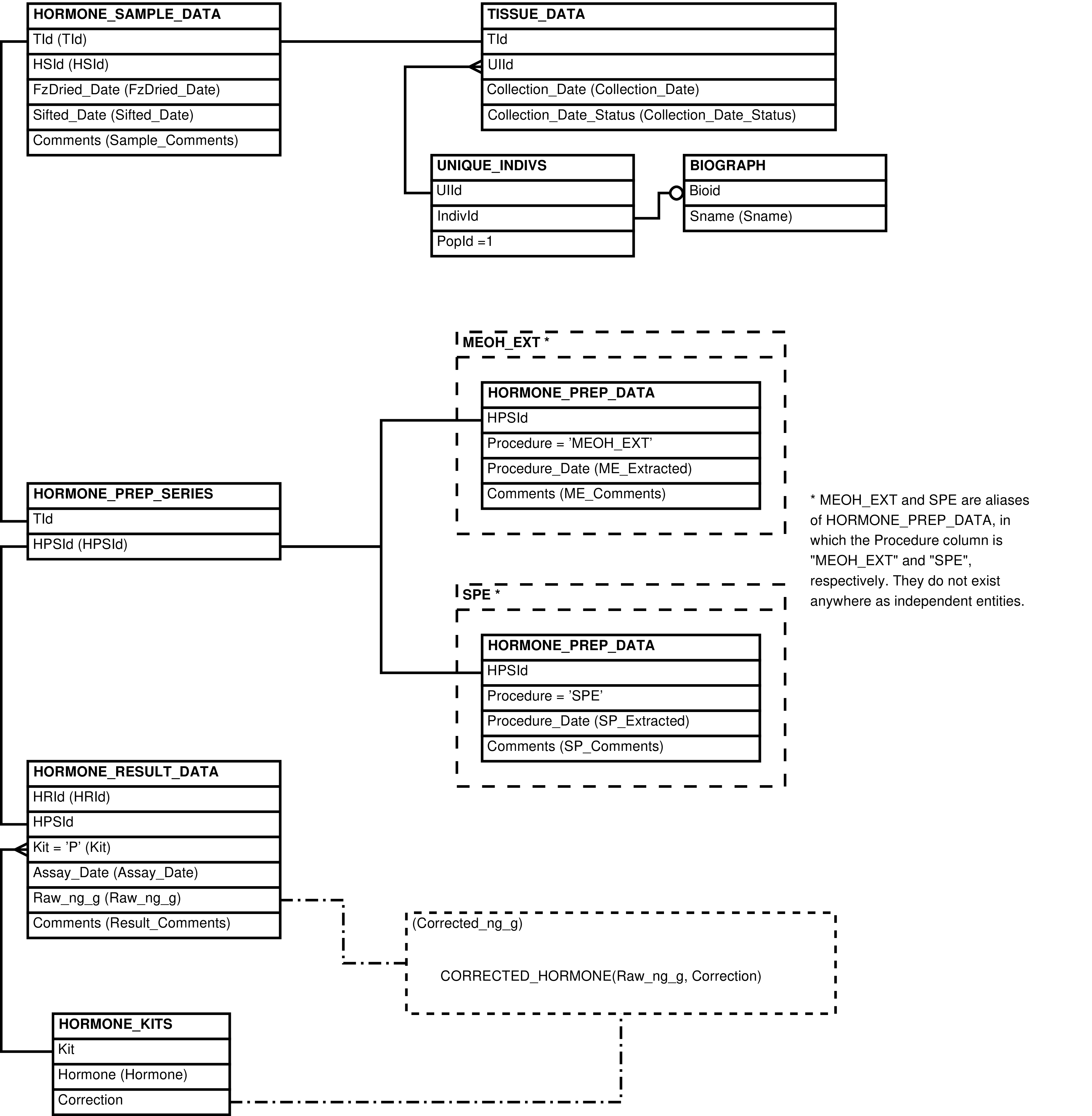
Contains one row for every sample whose progesterone
concentration has been measured by any kit with a known
correction factor. Results from kits whose HORMONE_KITS.Correction is NULL are
omitted.
Tip
Use this view to see progesterone concentration in all the hormone samples. It joins all the pertinent tables together to gather information, and omits results that are not considered reliable.
This view includes a "Hormone" column that indicates
which hormone was measured in each assay. By definition, this
column will be P in every
row, so it may seem odd to include the row at all. The column
is retained as a courtesy to users, especially for those who
might unify the rows from this view with rows of other,
similar views (e.g. ESTROGENS, GLUCOCORTICOIDS, etc.).
Figure 6.36. Query Defining the PROGESTERONES View
SELECT hormone_sample_data.tid
, hormone_prep_series.hpsid
, hormone_result_data.hrid
, hormone_sample_data.hsid
, biograph.sname
, tissue_data.collection_date
, tissue_data.collection_date_status AS collection_date_status
, hormone_sample_data.fzdried_date AS fzdried_date
, hormone_sample_data.sifted_date AS sifted_date
, meoh_ext.procedure_date AS me_extracted
, spe.procedure_date AS sp_extracted
, hormone_result_data.raw_ng_g AS raw_ng_g
, corrected_hormone(hormone_result_data.raw_ng_g, hormone_kits.correction) AS corrected_ng_g
, hormone_result_data.assay_date
, hormone_kits.hormone AS hormone
, hormone_result_data.kit AS kit
, hormone_sample_data.comments AS sample_comments
, hormone_result_data.comments AS result_comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ
JOIN hormone_prep_series
ON hormone_prep_series.tid = hormone_sample_data.tid
JOIN hormone_result_data
ON hormone_result_data.hpsid = hormone_prep_series.hpsid
JOIN hormone_kits
ON hormone_kits.kit = hormone_result_data.kit
AND hormone_kits.correction IS NOT NULL
AND hormone_kits.hormone = 'P'
LEFT JOIN hormone_prep_data AS meoh_ext
ON meoh_ext.procedure = 'MEOH_EXT'
AND meoh_ext.hpsid = hormone_prep_series.hpsid
LEFT JOIN hormone_prep_data AS spe
ON spe.procedure = 'SPE'
AND spe.hpsid = hormone_prep_series.hpsid;
Table 6.17. Columns in the PROGESTERONES View
| Column | From | Description |
|---|---|---|
| TId | HORMONE_SAMPLE_DATA.TId | Identifier of the HORMONE_SAMPLE_DATA row and tissue sample |
| HPSId | HORMONE_PREP_SERIES.HPSId | Identifier of the prep series |
| HRId | HORMONE_RESULT_DATA.HRId | Identifier of the assay that generated this result |
| HSId | HORMONE_SAMPLE_DATA.HSId | User-generated identifier for the tissue sample |
| Sname | BIOGRAPH.Sname | Sname of the individual from whom this sample came |
| Collection_Date | TISSUE_DATA.Collection_Date | Date the tissue sample was collected |
| Collection_Date_Status | TISSUE_DATA.Collection_Date_Status | The status of this Collection_Date |
| FzDried_Date | HORMONE_SAMPLE_DATA.FzDried_Date | Date the sample was freeze-dried |
| Sifted_Date | HORMONE_SAMPLE_DATA.Sifted_Date | Date the freeze-dried sample was sifted |
| ME_Extracted | HORMONE_PREP_DATA.Procedure_Date | Date of methanol extraction, in prep for this result |
| SP_Extracted | HORMONE_PREP_DATA.Procedure_Date | Date of solid-phase extraction, in prep for this result |
| Raw_ng_g | HORMONE_RESULT_DATA.Raw_ng_g | The "raw" concentration determined in this assay |
| Corrected_ng_g | CORRECTED_HORMONE(HORMONE_RESULT_DATA.Raw_ng_g, HORMONE_KITS.Correction) | The corrected concentration, according to the related HORMONE_KITS.Correction |
| Assay_Date | HORMONE_RESULT_DATA.Assay_Date | Date of this assay |
| Hormone | HORMONE_KITS.Hormone | Code for the hormone whose concentration was assayed |
| Kit | HORMONE_RESULT_DATA.Kit | Code for the kit used in this assay |
| Sample_Comments | HORMONE_SAMPLE_DATA.Comments | Comments about the hormone sample |
| ME_Comments | HORMONE_PREP_DATA.Comments | Comments about the methanol extraction |
| SP_Comments | HORMONE_PREP_DATA.Comments | Comments about the solid-phase extraction |
| Result_Comments | HORMONE_RESULT_DATA.Comments | Comments about the assay |
Contains one row for every sample whose testosterone
concentration has been measured by any kit with a known
correction factor. Results from kits whose HORMONE_KITS.Correction is NULL are
omitted.
Tip
Use this view to see testosterone concentration in all the hormone samples. It joins all the pertinent tables together to gather information, and omits results that are not considered reliable.
This view includes a "Hormone" column that indicates
which hormone was measured in each assay. By definition, this
column will be T in every
row, so it may seem odd to include the row at all. The column
is retained as a courtesy to users, especially for those who
might unify the rows from this view with rows of other,
similar views (e.g. ESTROGENS, GLUCOCORTICOIDS, etc.).
Figure 6.38. Query Defining the TESTOSTERONES View
SELECT hormone_sample_data.tid
, hormone_prep_series.hpsid
, hormone_result_data.hrid
, hormone_sample_data.hsid
, biograph.sname
, tissue_data.collection_date
, tissue_data.collection_date_status AS collection_date_status
, hormone_sample_data.fzdried_date AS fzdried_date
, hormone_sample_data.sifted_date AS sifted_date
, meoh_ext.procedure_date AS me_extracted
, spe.procedure_date AS sp_extracted
, hormone_result_data.raw_ng_g AS raw_ng_g
, corrected_hormone(hormone_result_data.raw_ng_g, hormone_kits.correction) AS corrected_ng_g
, hormone_result_data.assay_date
, hormone_kits.hormone AS hormone
, hormone_result_data.kit AS kit
, hormone_sample_data.comments AS sample_comments
, hormone_result_data.comments AS result_comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ
JOIN hormone_prep_series
ON hormone_prep_series.tid = hormone_sample_data.tid
JOIN hormone_result_data
ON hormone_result_data.hpsid = hormone_prep_series.hpsid
JOIN hormone_kits
ON hormone_kits.kit = hormone_result_data.kit
AND hormone_kits.correction IS NOT NULL
AND hormone_kits.hormone = 'T'
LEFT JOIN hormone_prep_data AS meoh_ext
ON meoh_ext.procedure = 'MEOH_EXT'
AND meoh_ext.hpsid = hormone_prep_series.hpsid
LEFT JOIN hormone_prep_data AS spe
ON spe.procedure = 'SPE'
AND spe.hpsid = hormone_prep_series.hpsid;
Table 6.18. Columns in the TESTOSTERONES View
| Column | From | Description |
|---|---|---|
| TId | HORMONE_SAMPLE_DATA.TId | Identifier of the HORMONE_SAMPLE_DATA row and tissue sample |
| HPSId | HORMONE_PREP_SERIES.HPSId | Identifier of the prep series |
| HRId | HORMONE_RESULT_DATA.HRId | Identifier of the assay that generated this result |
| HSId | HORMONE_SAMPLE_DATA.HSId | User-generated identifier for the tissue sample |
| Sname | BIOGRAPH.Sname | Sname of the individual from whom this sample came |
| Collection_Date | TISSUE_DATA.Collection_Date | Date the tissue sample was collected |
| Collection_Date_Status | TISSUE_DATA.Collection_Date_Status | The status of this Collection_Date |
| FzDried_Date | HORMONE_SAMPLE_DATA.FzDried_Date | Date the sample was freeze-dried |
| Sifted_Date | HORMONE_SAMPLE_DATA.Sifted_Date | Date the freeze-dried sample was sifted |
| ME_Extracted | HORMONE_PREP_DATA.Procedure_Date | Date of methanol extraction, in prep for this result |
| SP_Extracted | HORMONE_PREP_DATA.Procedure_Date | Date of solid-phase extraction, in prep for this result |
| Raw_ng_g | HORMONE_RESULT_DATA.Raw_ng_g | The "raw" concentration determined in this assay |
| Corrected_ng_g | CORRECTED_HORMONE(HORMONE_RESULT_DATA.Raw_ng_g, HORMONE_KITS.Correction) | The corrected concentration, according to the related HORMONE_KITS.Correction |
| Assay_Date | HORMONE_RESULT_DATA.Assay_Date | Date of this assay |
| Hormone | HORMONE_KITS.Hormone | Code for the hormone whose concentration was assayed |
| Kit | HORMONE_RESULT_DATA.Kit | Code for the kit used in this assay |
| Sample_Comments | HORMONE_SAMPLE_DATA.Comments | Comments about the hormone sample |
| ME_Comments | HORMONE_PREP_DATA.Comments | Comments about the methanol extraction |
| SP_Comments | HORMONE_PREP_DATA.Comments | Comments about the solid-phase extraction |
| Result_Comments | HORMONE_RESULT_DATA.Comments | Comments about the assay |
Contains one row for every sample whose thyroid hormone
concentration has been measured by any kit with a known
correction factor. Results from kits whose HORMONE_KITS.Correction is NULL are
omitted.
Tip
Use this view to see thyroid hormone concentration in all the hormone samples. It joins all the pertinent tables together to gather information, and omits results that are not considered reliable.
This view includes a "Hormone" column that indicates
which hormone was measured in each assay. By definition, this
column will be TH in every
row, so it may seem odd to include the row at all. The column
is retained as a courtesy to users, especially for those who
might unify the rows from this view with rows of other,
similar views (e.g. ESTROGENS, GLUCOCORTICOIDS, etc.).
Figure 6.40. Query Defining the THYROID_HORMONES View
SELECT hormone_sample_data.tid
, hormone_prep_series.hpsid
, hormone_result_data.hrid
, hormone_sample_data.hsid
, biograph.sname
, tissue_data.collection_date
, tissue_data.collection_date_status AS collection_date_status
, hormone_sample_data.fzdried_date AS fzdried_date
, hormone_sample_data.sifted_date AS sifted_date
, etoh_ext.procedure_date AS et_extracted
, hormone_result_data.raw_ng_g AS raw_ng_g
, corrected_hormone(hormone_result_data.raw_ng_g, hormone_kits.correction) AS corrected_ng_g
, hormone_result_data.assay_date
, hormone_kits.hormone AS hormone
, hormone_result_data.kit AS kit
, hormone_sample_data.comments AS sample_comments
, hormone_result_data.comments AS result_comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ
JOIN hormone_prep_series
ON hormone_prep_series.tid = hormone_sample_data.tid
JOIN hormone_result_data
ON hormone_result_data.hpsid = hormone_prep_series.hpsid
JOIN hormone_kits
ON hormone_kits.kit = hormone_result_data.kit
AND hormone_kits.correction IS NOT NULL
AND hormone_kits.hormone = 'TH'
LEFT JOIN hormone_prep_data AS etoh_ext
ON etoh_ext.procedure = 'ETOH_EXT'
AND etoh_ext.hpsid = hormone_prep_series.hpsid;
Table 6.19. Columns in the THYROID_HORMONES View
| Column | From | Description |
|---|---|---|
| TId | HORMONE_SAMPLE_DATA.TId | Identifier of the HORMONE_SAMPLE_DATA row and tissue sample |
| HPSId | HORMONE_PREP_SERIES.HPSId | Identifier of the prep series |
| HRId | HORMONE_RESULT_DATA.HRId | Identifier of the assay that generated this result |
| HSId | HORMONE_SAMPLE_DATA.HSId | User-generated identifier for the tissue sample |
| Sname | BIOGRAPH.Sname | Sname of the individual from whom this sample came |
| Collection_Date | TISSUE_DATA.Collection_Date | Date the tissue sample was collected |
| Collection_Date_Status | TISSUE_DATA.Collection_Date_Status | The status of this Collection_Date |
| FzDried_Date | HORMONE_SAMPLE_DATA.FzDried_Date | Date the sample was freeze-dried |
| Sifted_Date | HORMONE_SAMPLE_DATA.Sifted_Date | Date the freeze-dried sample was sifted |
| ET_Extracted | HORMONE_PREP_DATA.Procedure_Date | Date of ethanol extraction, in prep for this result |
| Raw_ng_g | HORMONE_RESULT_DATA.Raw_ng_g | The "raw" concentration determined in this assay |
| Corrected_ng_g | CORRECTED_HORMONE(HORMONE_RESULT_DATA.Raw_ng_g, HORMONE_KITS.Correction) | The corrected concentration, according to the related HORMONE_KITS.Correction |
| Assay_Date | HORMONE_RESULT_DATA.Assay_Date | Date of this assay |
| Hormone | HORMONE_KITS.Hormone | Code for the hormone whose concentration was assayed |
| Kit | HORMONE_RESULT_DATA.Kit | Code for the kit used in this assay |
| Sample_Comments | HORMONE_SAMPLE_DATA.Comments | Comments about the hormone sample |
| EE_Comments | HORMONE_PREP_DATA.Comments | Comments about the ethanol extraction |
| Result_Comments | HORMONE_RESULT_DATA.Comments | Comments about the assay |
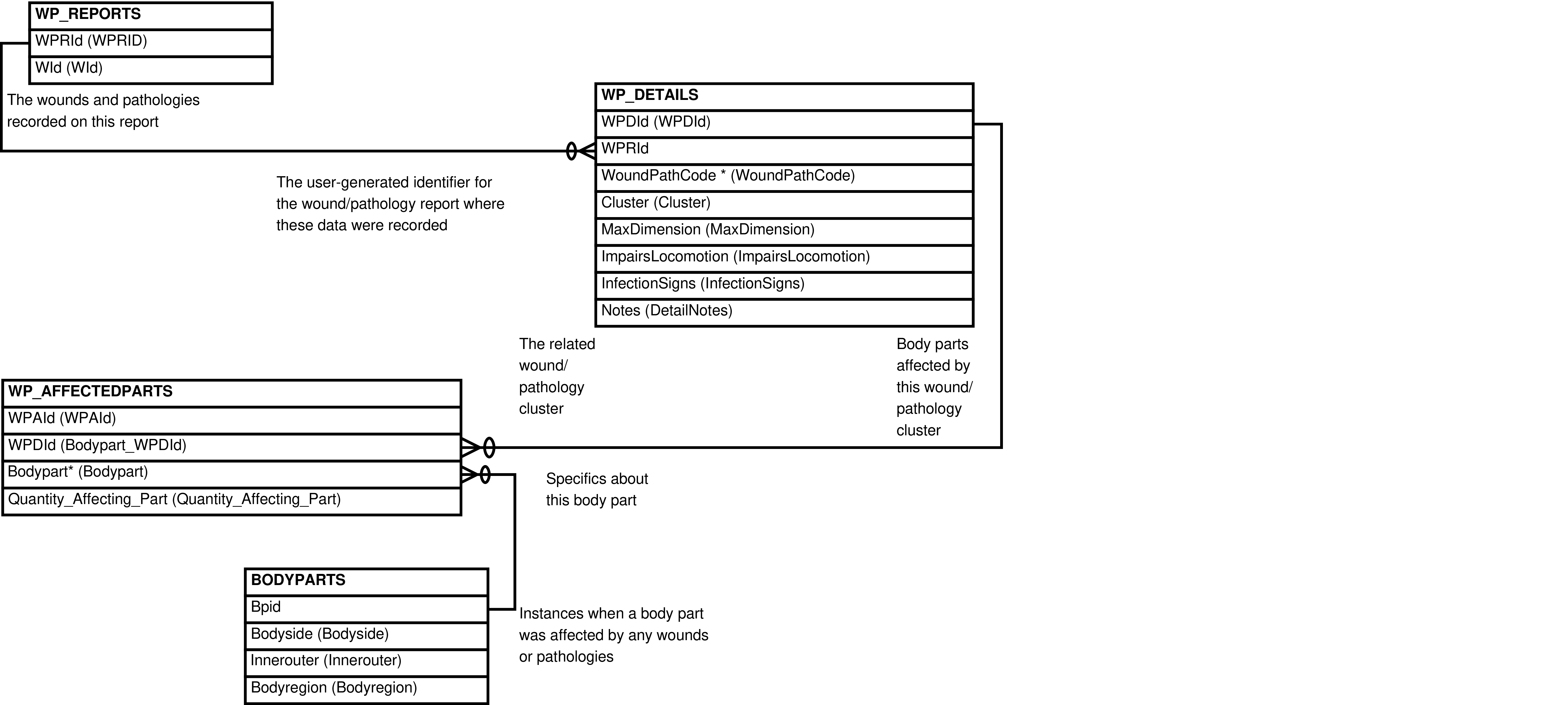
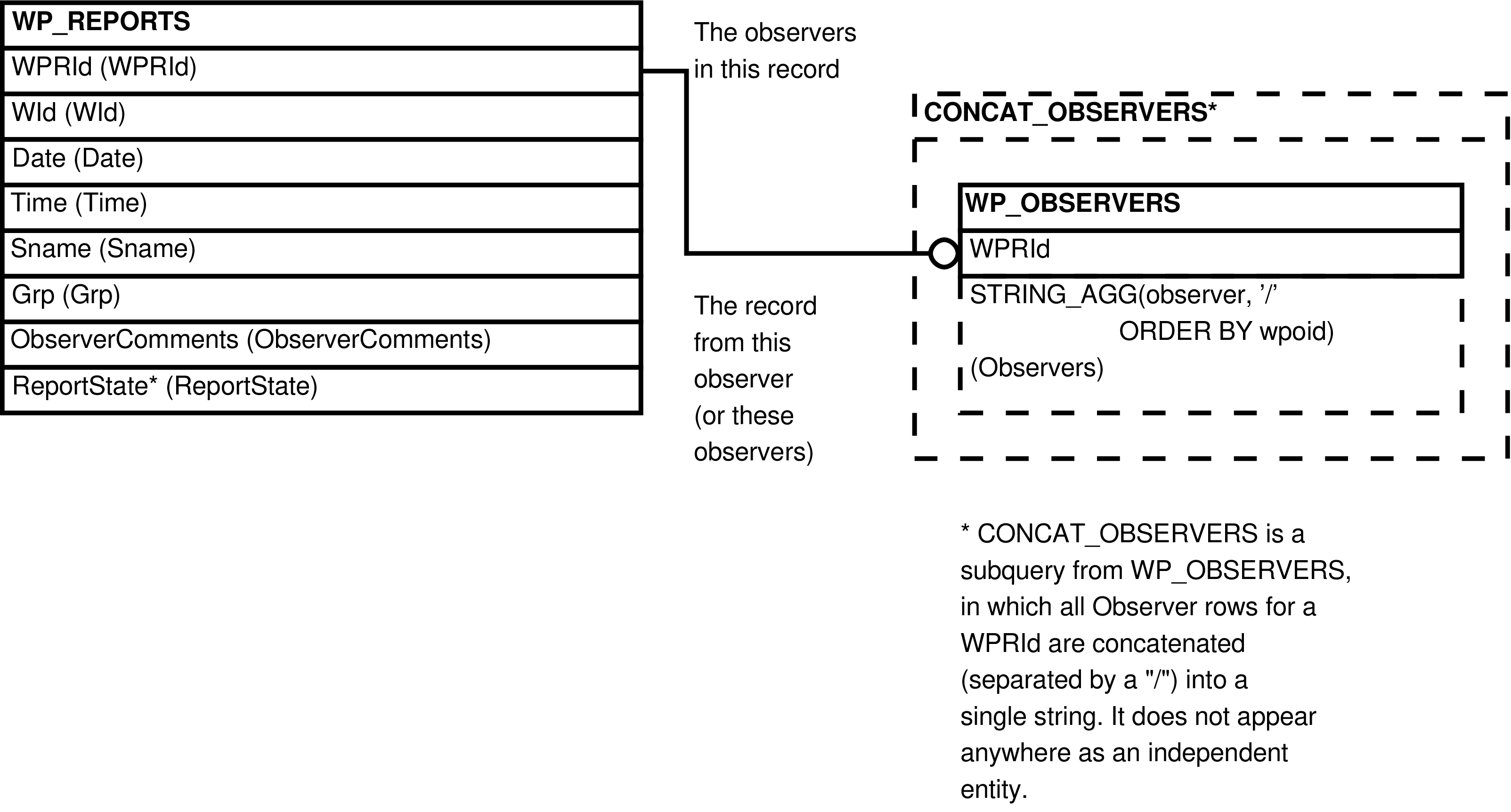
This view is intended to be the main place to visualize the wounds/pathologies data without heal updates. It contains one row for every body part affected in a wound/pathology cluster, including all related columns from WP_REPORTS, WP_DETAILS, WP_AFFECTEDPARTS and BODYPARTS, and a single column of concatenated observers from WP_OBSERVERS.
Tip
Use this view instead of the individual component tables when selecting wounds/pathologies data and when heal updates needn't be included.
Figure 6.42. Query Defining the WOUNDSPATHOLOGIES View
WITH concat_observers AS (SELECT wprid
, string_agg(observer, '/' ORDER BY wpoid) as observers
FROM wp_observers
GROUP BY wprid)
SELECT wp_reports.wprid AS wprid
, wp_reports.wid AS wid
, wp_reports.date AS reportdate
, wp_reports.time AS reporttime
, concat_observers.observers AS observers
, wp_reports.sname AS sname
, wp_reports.grp AS grp
, wp_reports.observercomments AS observercomments
, wp_reports.reportstate AS reportstate
, wp_details.wpdid AS wpdid
, wp_details.woundpathcode AS woundpathcode
, wp_details.cluster AS cluster
, wp_details.maxdimension AS maxdimension
, wp_details.impairslocomotion AS impairslocomotion
, wp_details.infectionsigns AS infectionsigns
, wp_details.notes AS detailnotes
, wp_affectedparts.wpaid AS wpaid
, wp_affectedparts.bodypart AS bodypart
, bodyparts.bodyside AS bodyside
, bodyparts.innerouter AS innerouter
, bodyparts.bodyregion AS bodyregion
, wp_affectedparts.quantity_affecting_part AS quantity_affecting_part
FROM wp_reports
LEFT JOIN concat_observers
ON concat_observers.wprid = wp_reports.wprid
LEFT JOIN wp_details
ON wp_details.wprid = wp_reports.wprid
LEFT JOIN wp_affectedparts
ON wp_affectedparts.wpdid = wp_details.wpdid
LEFT JOIN bodyparts
ON bodyparts.bpid = wp_affectedparts.bodypart;
Table 6.20. Columns in the WOUNDSPATHOLOGIES View
| Column | From | Description |
|---|---|---|
| WPRId | WP_REPORTS.WPRId | Identifier for the report. |
| WId | WP_REPORTS.WId | User-generated identifier for the report. |
| ReportDate | WP_REPORTS.Date | The date that the report was created. |
| ReportTime | WP_REPORTS.Time | The time that the report was created. |
| Observers | WP_OBSERVERS.Observer | All of the report's observers, concatenated
together and separated by a
"/". If no
observers, then NULL. |
| Sname | WP_REPORTS.Sname | The sname of the affected individual. |
| Grp | WP_REPORTS.Grp | The group of the individual in the report, according to the observer(s). |
| ObserverComments | WP_REPORTS.ObserverComments | Notes or comments from the observer(s) about the report. |
| ReportState | WP_REPORTS.ReportState | Status of the report. |
| WPDId | WP_DETAILS.WPDId | Identifier for the wound/pathology cluster. |
| WoundPathCode | WP_DETAILS.WoundPathCode | Code indicating the wound/pathology type for the cluster. |
| Cluster | WP_DETAILS.Cluster | The wound/pathology cluster identifier. |
| MaxDimension | WP_DETAILS.MaxDimension | The highest observed length, height, depth, etc. (as applicable), in cm, of all wounds/pathologies in the cluster. |
| ImpairsLocomotion | WP_DETAILS.ImpairsLocomotion | Boolean indicating whether or not the wound/pathology cluster impairs the individual's locomotion. |
| InfectionSigns | WP_DETAILS.InfectionSigns | Boolean indicating whether or not the wound/pathology cluster includes signs of an infection. |
| DetailNotes | WP_DETAILS.Notes | Textual comments or notes about the cluster. |
| WPAId | WP_AFFECTEDPARTS.WPAId | Identifier for the affected body part in the wound/pathology cluster. |
| Bodypart | WP_AFFECTEDPARTS.Bodypart | Unique identifier for the body part. |
| Bodyside | BODYPARTS.Bodyside | Code indicating the side of the body on which the affected part is located. |
| Innerouter | BODYPARTS.Innerouter | Code indicating if the affected body part is on the inner or outer side of the body part. |
| Bodyregion | BODYPARTS.Bodyregion | Code indicating the region on the body of the affected body part. |
| Quantity_Affecting_Part | WP_AFFECTEDPARTS.Quantity_Affecting_Part | The number of wounds/pathologies described in the cluster affecting this body part. |
Contains one row for every row in WP_AFFECTEDPARTS, with related identifier columns from WP_REPORTS and related data from the WP_DETAILS and BODYPARTS tables.
The intended purpose of this view is for uploading data into WP_DETAILS and WP_AFFECTEDPARTS. It may also be useful for querying/accessing the data.
Figure 6.44. Query Defining the WP_DETAILS_AFFECTEDPARTS View
SELECT wp_details.wpdid AS wpdid
, wp_reports.wprid AS wprid
, wp_reports.wid AS wid
, wp_details.woundpathcode AS woundpathcode
, wp_details.cluster AS cluster
, wp_details.maxdimension AS maxdimension
, wp_details.impairslocomotion AS impairslocomotion
, wp_details.infectionsigns AS infectionsigns
, wp_details.notes AS detailnotes
, wp_affectedparts.wpaid AS wpaid
, wp_affectedparts.wpdid AS bodypart_wpdid
, wp_affectedparts.bodypart AS bodypart
, bodyparts.bodyside AS bodyside
, bodyparts.innerouter AS innerouter
, bodyparts.bodyregion AS bodyregion
, wp_affectedparts.quantity_affecting_part AS quantity_affecting_part
FROM wp_reports
JOIN wp_details
ON wp_details.wprid = wp_reports.wprid
LEFT JOIN wp_affectedparts
ON wp_affectedparts.wpdid = wp_details.wpdid
LEFT JOIN bodyparts
ON bodyparts.bpid = wp_affectedparts.bodypart;
Table 6.21. Columns in the WP_DETAILS_AFFECTEDPARTS View
| Column | From | Description |
|---|---|---|
| WPDId | WP_DETAILS.WPDId | Identifier for the wound/pathology cluster. |
| WPRId | WP_REPORTS.WPRId | Identifier for the report in which these wounds/pathologies were recorded. |
| WId | WP_REPORTS.WId | User-generated identifier for the report in which these wounds/pathologies were recorded. |
| WoundPathCode | WP_DETAILS.WoundPathCode | Code indicating the wound/pathology type. |
| Cluster | WP_DETAILS.Cluster | The wound/pathology cluster identifier. |
| MaxDimension | WP_DETAILS.MaxDimension | The highest observed length, height, depth, etc. (as applicable), in cm, of all wounds/pathologies in this cluster. |
| ImpairsLocomotion | WP_DETAILS.ImpairsLocomotion | Boolean indicating whether or not this wound/pathology cluster impairs the individual's locomotion. |
| InfectionSigns | WP_DETAILS.InfectionSigns | Boolean indicating whether or not this wound/pathology cluster includes signs of an infection. |
| DetailNotes | WP_DETAILS.Notes | Textual comments or notes about this cluster. |
| WPAId | WP_AFFECTEDPARTS.WPAId | Identifier for the affected body part in this
wound/pathology cluster. If there are no related
rows in WP_AFFECTEDPARTS, then
NULL. |
| Bodypart_WPDId | WP_AFFECTEDPARTS.WPDId | Identifier for the wound/pathology cluster,
from WP_AFFECTEDPARTS. When
selecting data, this will always equal the WPDId
column. This column is included to allow the ability
to change the WP_AFFECTEDPARTS.WPDId with an
UPDATE command. If there are no
related rows in WP_AFFECTEDPARTS,
then NULL. |
| Bodypart | WP_AFFECTEDPARTS.Bodypart, BODYPARTS.Bpid | Unique identifier for the body part. If there
are no related rows in WP_AFFECTEDPARTS, then NULL. |
| Bodyside | BODYPARTS.Bodyside | Code indicating the side of the body on which
the affected part is located. If there are no
related rows in WP_AFFECTEDPARTS,
then NULL. |
| Innerouter | BODYPARTS.Innerouter | Code indicating if the affected body part is
on the inner or outer side of the body part. If
there are no related rows in WP_AFFECTEDPARTS, then NULL. |
| Bodyregion | BODYPARTS.Bodyregion | Code indicating the region on the body of the
affected body part. If there are no related rows in
WP_AFFECTEDPARTS, then
NULL. |
| Quantity_Affecting_Part | WP_AFFECTEDPARTS.Quantity_Affecting_Part | The number of wounds/pathologies described in
the cluster affecting this body part. If there are
no related rows in WP_AFFECTEDPARTS, then NULL. |
At least one of the WPRId and WId columns cannot be
NULL; these values are used to determine the related WP_DETAILS.WPRId. If both are provided, that
pair must already exist as a WPRId-WId pair in WP_REPORTS.
There
must be enough body part information provided to identify a
single body part code for the WP_AFFECTEDPARTS.Bodypart column. This means
that the provided Bodypart column must not be NULL or the
provided Bodyside-Innerouter-Bodyregion must be identical to
those of a single row in BODYPARTS. If
the Bodypart is not NULL and one or more of the Bodyside,
Innerouter, or Bodyregion columns is also not NULL, it is
an error if any of the provided Bodyside, Innerouter, or
Bodyregion values does not match their related columns in
BODYPARTS for the provided Bodypart (Bpid).
Inserting a row into WP_DETAILS_AFFECTEDPARTS inserts a row into WP_DETAILS and WP_AFFECTEDPARTS, as described below.
Like their related columns in WP_DETAILS, the WoundPathCode, Cluster,
ImpairsLocomotion, and InfectionSigns columns cannot be
NULL. When there is already a WP_DETAILS row with the provided
WoundPathCode, Cluster, MaxDimension, ImpairsLocomotion,
InfectionSigns, DetailNotes (Notes), and either WPRId or related
WId, a new WP_DETAILS row is not added.
These values are still used to determine the correct WPDId
to use when inserting data into WP_AFFECTEDPARTS.
When Bodypart_WPDId is not provided, new WP_AFFECTEDPARTS rows are inserted using the WPDId of the related WP_DETAILS row. If a Bodypart_WPDId is provided, it must equal the related WPDId from WP_DETAILS, whether or not WPDId is provided.
The new WP_AFFECTEDPARTS.Bodypart value is determined
with the provided body part columns, as described above.
When the Bodypart column is NULL, it is an error if one
or more of the Bodyside, Innerouter, or Bodyregion columns
is also NULL.
Updating a row in WP_DETAILS_AFFECTEDPARTS updates the underlying columns in WP_DETAILS and WP_AFFECTEDPARTS, as expected.
Tip
To update the WPDId in a WP_AFFECTEDPARTS row, update the Bodypart_WPDId column, not the WPDId. The former exists explicitly for this purpose, while the latter refers to the WPDId column in WP_DETAILS, which cannot be changed.
Deleting a row in WP_DETAILS_AFFECTEDPARTS deletes the underlying rows in WP_DETAILS and in WP_AFFECTEDPARTS.
DELETE commands in this view
remove the WP_DETAILS row, and all
related WP_AFFECTEDPARTS rows are
deleted concomitantly. It is not possible to
only remove row(s) from WP_AFFECTEDPARTS when deleting from this
view.
Tip
To remove WP_AFFECTEDPARTS rows without deleting the related WP_DETAILS row, don't use this view. You should manually delete the rows from the WP_AFFECTEDPARTS table.
Contains one row for every row in WP_HEALUPDATES, with related columns from WP_REPORTS, WP_DETAILS, WP_AFFECTEDPARTS, and BODYPARTS, and an "Observers" column concatenating all related WP_OBSERVERS.Observer values together. Whether or not a particular table's rows are "related" depends somewhat on the specificity of the heal update, as discussed below.
Although the relationship between WP_REPORTS, WP_OBSERVERS, WP_DETAILS, WP_AFFECTEDPARTS,
and BODYPARTS rows is unambiguous, the
relationship between them and a particular heal update may not
be so straightforward. For example, a heal update for a
particular cluster (when the WP_HEALUPDATES.WPDId is not NULL) might in
reality apply to one or all of that cluster's affected body
parts, but the update's being recorded for the cluster
indicates that it is unspecified or unknown which specific
body parts have healed. Users may decide on their own to make
assumptions about which body parts are included in such an
update, but it would be misleading for this view to join them
together and imply more specificity than is actually known.
To prevent such false implications of specificity, this view
leaves NULL any columns that are more specific than what is
indicated in the WP_HEALUPDATES
row. Specifically: when the update is for a report (the WPRId is not NULL), the related
values from WP_REPORTS will be returned,
while those from WP_DETAILS, WP_AFFECTEDPARTS, and BODYPARTS
will be NULL. When the update is for a cluster (the WPDId is not NULL), the related
values from WP_REPORTS and WP_DETAILS will be returned, but those from WP_AFFECTEDPARTS and BODYPARTS
will be NULL. An update for an affected body part (the WPAId is not NULL) will return
the related values from all the aforementioned tables.
Regardless of update specificity, the concatenated
"Observers" column will always be included. It will be NULL
only when there are no observers recorded for the related
report in WP_OBSERVERS.
Caution
Many of the tables in this view have a "one-to-many" relationship with each other. Because of this, some normally-unique values may appear to be duplicated across multiple rows. Remember, only WP_HEALS rows are truly unique in this view.
Tip
Use this view when selecting wounds/pathologies data that include the heal updates. (Use this instead of the WP_HEALUPDATES table.) This view presents the data in a format more hospitable for humans to read, and performs the somewhat-tricky task of joining the different ID columns (WPRId, WPDId, and WPAId) to their respective tables.
Figure 6.46. Query Defining the WP_HEALS View
WITH concat_observers AS (SELECT wprid
, string_agg(observer, '/' ORDER BY wpoid) as observers
FROM wp_observers
GROUP BY wprid)
SELECT wp_reports.wprid AS wprid
, wp_reports.wid AS wid
, wp_reports.date AS reportdate
, wp_reports.time AS reporttime
, concat_observers.observers AS observers
, wp_reports.sname AS sname
, wp_reports.grp AS grp
, wp_reports.observercomments AS observercomments
, wp_reports.reportstate AS reportstate
, wp_details.wpdid AS wpdid
, wp_details.woundpathcode AS woundpathcode
, wp_details.cluster AS cluster
, wp_details.maxdimension AS maxdimension
, wp_details.impairslocomotion AS impairslocomotion
, wp_details.infectionsigns AS infectionsigns
, wp_details.notes AS detailnotes
, wp_affectedparts.wpaid AS wpaid
, wp_affectedparts.bodypart AS bodypart
, bodyparts.bodyside AS bodyside
, bodyparts.innerouter AS innerouter
, bodyparts.bodyregion AS bodyregion
, wp_affectedparts.quantity_affecting_part AS quantity_affecting_part
, wp_healupdates.wphid AS wphid
, wp_healupdates.date AS healdate
, wp_healupdates.healstatus AS healstatus
, wp_healupdates.notes AS healnotes
FROM wp_healupdates
LEFT JOIN wp_affectedparts
ON wp_affectedparts.wpaid = wp_healupdates.wpaid
LEFT JOIN bodyparts
ON bodyparts.bpid = wp_affectedparts.bodypart
LEFT JOIN wp_details
ON wp_details.wpdid = COALESCE(wp_affectedparts.wpdid, wp_healupdates.wpdid)
LEFT JOIN wp_reports
ON wp_reports.wprid = COALESCE(wp_details.wprid, wp_healupdates.wprid)
LEFT JOIN concat_observers
ON concat_observers.wprid = wp_reports.wprid;
Figure 6.48. Entity Relationship Diagram of the WP_HEALS View for rows with an update to a wound/pathology report

|
Figure 6.49. Entity Relationship Diagram of the WP_HEALS View for rows with an update to a wound/pathology cluster

|
Figure 6.50. Entity Relationship Diagram of the WP_HEALS View for rows with an update to an affected body part
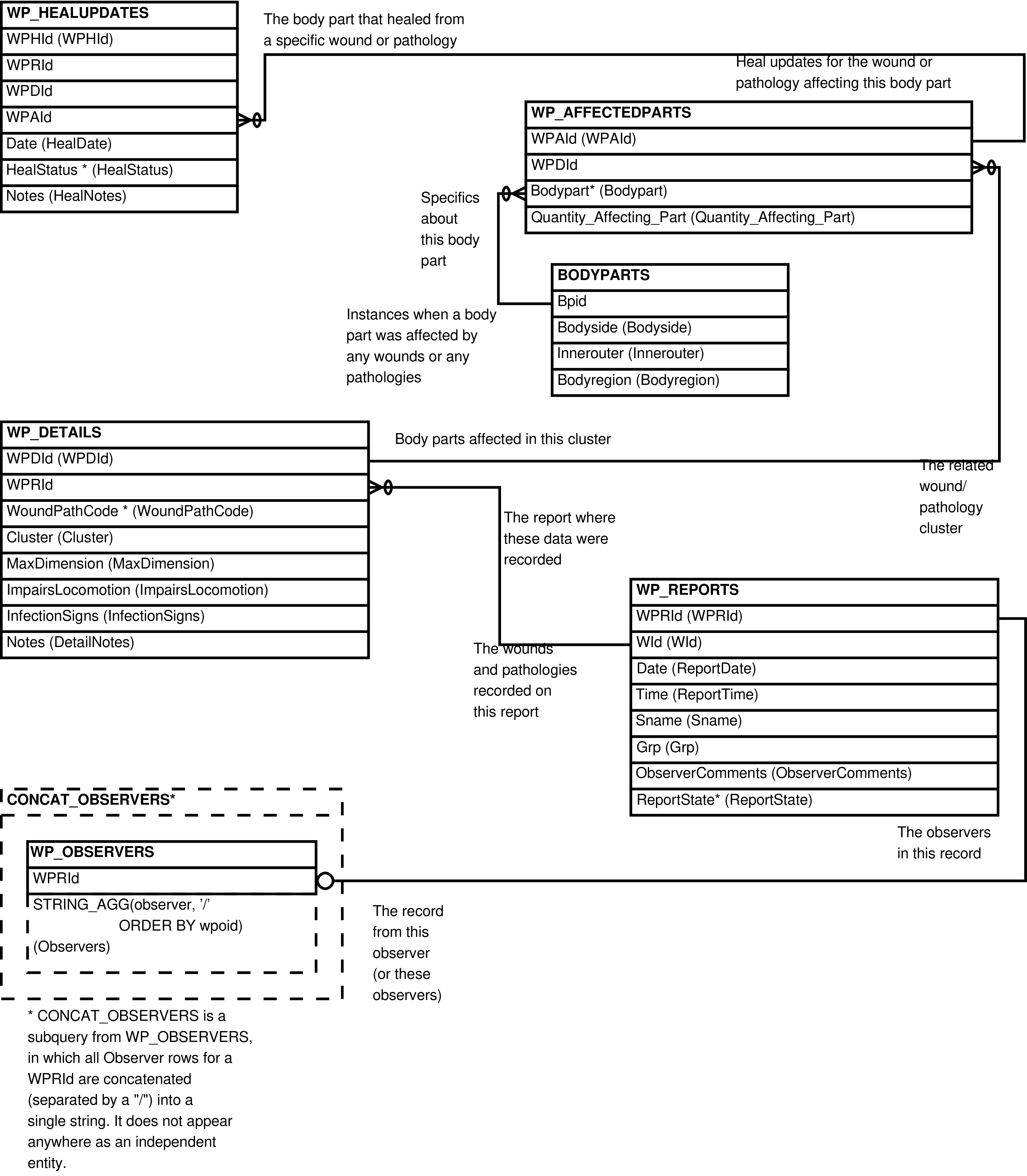
|
Table 6.22. Columns in the WP_HEALS View
| Column | From | Description |
|---|---|---|
| WPRId | WP_REPORTS.WPRId | Identifier for the report. |
| WId | WP_REPORTS.WId | User-generated identifier for the report. |
| ReportDate | WP_REPORTS.Date | The date that the report was created. |
| ReportTime | WP_REPORTS.Time | The time that the report was created. |
| Observers | WP_OBSERVERS.Observer | All of the report's observers, concatenated
together and separated by a
"/". If no
observers, then NULL. |
| Sname | WP_REPORTS.Sname | The sname of the affected individual. |
| Grp | WP_REPORTS.Grp | The group of the individual in the report, according to the observer(s). |
| ObserverComments | WP_REPORTS.ObserverComments | Notes or comments from the observer(s) about the report. |
| ReportState | WP_REPORTS.ReportState | Status of the report. |
| WPDId | WP_DETAILS.WPDId | Identifier for the wound/pathology
cluster. NULL if this heal update is only for the
report. |
| WoundPathCode | WP_DETAILS.WoundPathCode | Code indicating the wound/pathology type for
the cluster. NULL if this heal update is only for
the report. |
| Cluster | WP_DETAILS.Cluster | The wound/pathology cluster
identifier. NULL if this heal update is only for
the report. |
| MaxDimension | WP_DETAILS.MaxDimension | The highest observed length, height, depth,
etc. (as applicable), in cm, of all
wounds/pathologies in the cluster. NULL if this
heal update is only for the report. |
| ImpairsLocomotion | WP_DETAILS.ImpairsLocomotion | Boolean indicating whether or not the
wound/pathology cluster impairs the individual's
locomotion. NULL if this heal update is only for
the report. |
| InfectionSigns | WP_DETAILS.InfectionSigns | Boolean indicating whether or not the
wound/pathology cluster includes signs of an
infection. NULL if this heal update is only for
the report. |
| DetailNotes | WP_DETAILS.Notes | Textual comments or notes about the
cluster. NULL if this heal update is only for the
report. |
| WPAId | WP_AFFECTEDPARTS.WPAId | Identifier for the affected body part in the
wound/pathology cluster. NULL if this heal update
is only for the report or the cluster. |
| Bodypart | WP_AFFECTEDPARTS.Bodypart | Unique identifier for the body part. NULL
if this heal update is only for the report or the
cluster. |
| Bodyside | BODYPARTS.Bodyside | Code indicating the side of the body on which
the affected part is located. NULL if this heal
update is only for the report or the
cluster. |
| Innerouter | BODYPARTS.Innerouter | Code indicating if the affected body part is
on the inner or outer side of the body part. NULL
if this heal update is only for the report or the
cluster. |
| Bodyregion | BODYPARTS.Bodyregion | Code indicating the region on the body of the
affected body part. NULL if this heal update is
only for the report or the cluster. |
| Quantity_Affecting_Part | WP_AFFECTEDPARTS.Quantity_Affecting_Part | The number of wounds/pathologies described in
the cluster affecting this body part. NULL if this
heal update is only for the report or the
cluster. |
| WPHId | WP_HEALUPDATES.WPHId | Identifier for the heal update. |
| HealDate | WP_HEALUPDATES.Date | Date of this heal update. |
| HealStatus | WP_HEALUPDATES.HealStatus | Code indicating how well the related wound/pathology has healed. |
| HealNotes | WP_HEALUPDATES.Notes | Textual notes about the healing (or lack thereof) in this update. |
Inserting a row into WP_HEALS inserts a row into WP_HEALUPDATES, as described below.
For each row inserted into WP_HEALUPDATES, the inserted WPRId, WPDId, or WPAId value is determined based on the values provided for the other columns in this view, as described below.
To insert a WP_HEALUPDATES row
updating a report (having a
non-NULL WPRId), the
provided data must be sufficient to uniquely identify a
single row in WP_REPORTS, and should not
include any information about clusters or affected body
parts. That is, the provided values in columns from WP_REPORTS (WPRId, WId, ReportDate,
ReportTime, Sname, Grp, ObserverComments, or ReportState)
and the "Observers" must altogether be associable with a
single report, and all the columns from WP_DETAILS (WPDId, WoundPathCode, Cluster,
MaxDimension, ImpairsLocomotion, InfectionSigns, and
DetailNotes), and both WP_AFFECTEDPARTS
and BODYPARTS (WPAId, Bodypart,
Bodyside, Innerouter, Bodyregion, and
Quantity_Affecting_Part) must all be NULL. It is not
necessary to provide all of the columns from WP_REPORTS or the Observers, just enough data
to uniquely identify the report. The WPRId of the designated WP_REPORTS row is inserted as the new WP_HEALUPDATES.WPRId.
To insert a WP_HEALUPDATES row
updating a cluster (having a
non-NULL WPDId), the
provided data must be sufficient to uniquely identify a
single row in WP_DETAILS, and should not
include any information about affected body parts. That
is, the provided values in columns from WP_DETAILS (WPDId, WoundPathCode, Cluster,
MaxDimension, ImpairsLocomotion, InfectionSigns, and
DetailNotes) and WP_REPORTS (WPRId, WId,
ReportDate, ReportTime, Sname, Grp, ObserverComments, and
ReportState), and the "Observers" must altogether be
associable with a single cluster and related report, and
all the columns from both WP_AFFECTEDPARTS and BODYPARTS (WPAId, Bodypart, Bodyside,
Innerouter, Bodyregion, and Quantity_Affecting_Part) must
all be NULL. It is not necessary to provide all of the
columns from WP_DETAILS or WP_REPORTS or the Observers, just enough data
to uniquely identify the cluster. The WPDId of the designated WP_DETAILS row is inserted as the new WP_HEALUPDATES.WPDId.
To insert a WP_HEALUPDATES row
updating an affected body part
(having a non-NULL WPAId), the provided data must
be sufficient to uniquely identify a single row in WP_AFFECTEDPARTS. That is, the provided
values must altogether be associable with a single body
part, related cluster, and related report. It is not
necessary to provide all of the columns from WP_AFFECTEDPARTS, WP_DETAILS, WP_REPORTS or
the "Observers", just enough data to uniquely identify the
affected body part. The WPAId of the designated WP_AFFECTEDPARTS row is inserted as the new
WP_HEALUPDATES.WPAId.
Each new WP_HEALUPDATES row is inserted with the provided HealDate, HealStatus, and HealNotes values.
Updating a row in WP_HEALS updates the underlying columns in WP_HEALUPDATES, as described below.
Updates to the HealDate, HealStatus, and HealNotes columns update their related columns in WP_HEALUPDATES, as expected. Updates to all other columns are prohibited[250].
Tip
To update the WPRId, WPDId, or WPAId columns in a WP_HEALUPDATES row, delete the WP_HEALUPDATES row and re-enter it with updated information.
Deleting a row in WP_HEALS deletes the underlying row in WP_HEALUPDATES. Related rows in WP_REPORTS, WP_DETAILS, WP_AFFECTEDPARTS, and BODYPARTS will be unaffected.
Contains one row for every row in WP_REPORTS. In addition to all of the columns
from WP_REPORTS, this view also has an
"Observers" column showing all related WP_OBSERVERS.Observer values (if any) concatenated
together (or NULL if there are no related values).
The intended purpose of this view is for uploading data into WP_REPORTS and WP_OBSERVERS, especially multiple observers for a single report. It may also be useful for querying/accessing the data.
When
uploading data with this view, it is an error if observer
initials cannot be unambiguously interpreted. In the
admittedly-unlikely event that there is an observer whose
initials legitimately include the separator character
"/", this observer's
initials cannot be inserted via this view.[251] In this case, the offending observer code must
be removed from the data, then manually inserted into WP_OBSERVERS.
Figure 6.51. Query Defining the WP_REPORTS_OBSERVERS View
WITH concat_observers AS (SELECT wprid
, string_agg(observer, '/' ORDER BY wpoid) as observers
FROM wp_observers
GROUP BY wprid)
SELECT wp_reports.wprid AS wprid
, wp_reports.wid AS wid
, wp_reports.date AS date
, wp_reports.time AS time
, concat_observers.observers AS observers
, wp_reports.sname AS sname
, wp_reports.grp AS grp
, wp_reports.observercomments AS observercomments
, wp_reports.reportstate AS reportstate
FROM wp_reports
LEFT JOIN concat_observers
ON concat_observers.wprid = wp_reports.wprid;
Table 6.23. Columns in the WP_REPORTS_OBSERVERS View
| Column | From | Description |
|---|---|---|
| WPRId | WP_REPORTS.WPRId | Identifier for the report. |
| WId | WP_REPORTS.WId | User-generated identifier for the report. |
| Date | WP_REPORTS.Date | The date that this report was created. |
| Time | WP_REPORTS.Time | The time that this report was created. |
| Observers | WP_OBSERVERS.Observer | All of this report's related observers,
concatenated together and separated by a
"/". If no
related observers, then NULL. |
| Sname | WP_REPORTS.Sname | The sname of the affected individual. |
| Grp | WP_REPORTS.Grp | The group of the individual in this report, according to the observer(s). |
| ObserverComments | WP_REPORTS.ObserverComments | Notes or comments from the observer(s) about this report. |
| ReportState | WP_REPORTS.ReportState | Status of the report. |
Inserting a row into WP_REPORTS_OBSERVERS inserts a row into WP_REPORTS and a number of rows into WP_OBSERVERS, as described below.
Like their related columns in WP_REPORTS, the WId, Date, Sname, Grp, and
ReportState columns cannot be NULL. When there is
already a WP_REPORTS row with the
provided WPRId, WId, ReportState, Date, Time, Sname, Grp,
and ObserverComments, a new WP_REPORTS
row is not added. These values are instead used to
determine the correct WPRId to use when inserting data
into WP_OBSERVERS.
For
each "/"-separated
observer provided in the Observers column, one row is
inserted into the WP_OBSERVERS table,
with the related WPRId. A
NULL Observers column is interpreted to mean that there
are no new rows to add to WP_OBSERVERS;
it does not result in a new WP_OBSERVERS row with a NULL Observer value.
Any observer initials provided that are already present for this WPRId in WP_OBSERVERS will not be added again.
Tip
To add new observers to a report that already has some observers recorded, you can insert a row that lists all the observers--already-present and not--or a row that only lists the newly-added observers.
Updating a row in WP_REPORTS_OBSERVERS updates the underlying row in WP_REPORTS as expected, and the underlying row(s) in WP_OBSERVERS as described below.
When an update doesn't actually change the Observers column, the related data in WP_OBSERVERS are unaffected. When the update does change the Observers column, all prior rows in WP_OBSERVERS are deleted, and new rows are inserted as described above.
Deleting a row in WP_REPORTS_OBSERVERS deletes the underlying row in WP_REPORTS and the underlying rows (if any) in WP_OBSERVERS.
DELETE commands in this view
remove the WP_REPORTS row, and all
related WP_OBSERVERS rows are
deleted concomitantly. It is not possible to
only remove row(s) from WP_OBSERVERS when deleting from this
view.
Tip
To remove any observers from a report without
deleting the related WP_REPORTS row,
use the UPDATE command in this view
(see above). Alternatively, skip the view altogether
and just delete the rows directly from the WP_OBSERVERS table.
[249] Normally, Series would not be enough to determine the correct HPSId, but is okay in this case because TId and/or HSId are also required.
[250] It is technically _possible_ to allow this view to accept updates to the other columns, and thus allow updates to the WPRId, WPDId, and WPAId columns in WP_HEALUPDATES. However, unless the user is willing to address most or all of the columns in this view in their update command, the validation needing to be written for these updates is prohibitively lengthy. The yield in utility does not seem worth the time investment to write such a thing, hence these kinds of updates are prohibited.
[251] It is possible to improve the algorithm used to
discern valid observer codes. This would reduce the need
for manual intervention on the part of the data manager at
the cost of increased complexity in the code. Given the
extreme UNlikelihood that there will ever be an observer
whose initials include a
"/", this improvement
seems unnecessary.