Technical Specifications for the Amboseli Baboon Project Data Management System
Anne Ndeti Hubbard
DocBook formattingKarl O. Pinc
DocBook formattingDocument generated: 2026-01-23 14:34:26.
Copyright Notices
Copyright (C) 2005-2023 Karl O. Pinc, Jeanne Altmann, Susan Alberts, Leah Gerber, Jake Gordon, The Meme Factory, Inc.
Except as otherwise noted permission is granted to copy, distribute and/or modify this document under the terms of the GNU Free Documentation License, Version 1.2 or any later version published by the Free Software Foundation; with no Invariant Sections, no Front-Cover Texts, and no Back-Cover Texts. A copy of the license is included in the section entitled “GNU Free Documentation License.”
Copyright (C) 1996-2011 The PostgreSQL Global Development Group
The appendix titled Database Transactions Explained is Copyright (C) 1996-2011 by the PostgreSQL Global Development Group, distributed under the terms of the license of the University of California below.
Permission to use, copy, modify, and distribute this software and its documentation for any purpose, without fee, and without a written agreement is hereby granted, provided that the above copyright notice and this paragraph and the following two paragraphs appear in all copies.
IN NO EVENT SHALL THE UNIVERSITY OF CALIFORNIA BE LIABLE TO ANY PARTY FOR DIRECT, INDIRECT, SPECIAL, INCIDENTAL, OR CONSEQUENTIAL DAMAGES, INCLUDING LOST PROFITS, ARISING OUT OF THE USE OF THIS SOFTWARE AND ITS DOCUMENTATION, EVEN IF THE UNIVERSITY OF CALIFORNIA HAS BEEN ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
THE UNIVERSITY OF CALIFORNIA SPECIFICALLY DISCLAIMS ANY WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE. THE SOFTWARE PROVIDED HEREUNDER IS ON AN "AS-IS" BASIS, AND THE UNIVERSITY OF CALIFORNIA HAS NO OBLIGATIONS TO PROVIDE MAINTENANCE, SUPPORT, UPDATES, ENHANCEMENTS, OR MODIFICATIONS.
March 2, 2005
We gratefully acknowledge the support of the National Science Foundation for the supporting the collection of the majority of the data stored in the database; in the past decade in particular we acknowledge support from IBN 9985910, IBN 0322613, IBN 0322781, BCS 0323553, BCS 0323596, DEB 0846286, DEB 0846532 and DEB 0919200. We are also very grateful for support from the National Institute of Aging (R01AG034513-01 and P01AG031719) and the Princeton Center for the Demography of Aging (P30AG024361). We also thank the Chicago Zoological Society, the Max Planck Institute for Demographic Research, the L.S.B. Leakey Foundation and the National Geographic Society for support at various times over the years. In addition, we thank the National Institute of Aging (R03-AG045459-01) for supporting recent work extending the database to incorporate genetic and genomic data.
Any opinions, findings, conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation, the National Institute of Aging, the Princeton Center for the Demography of Aging, the Chicago Zoological Society, the Max Planck Institute for Demographic Research, the L.S.B. Leakey Foundation, the National Geographic Society, or any other organization which has supplied support for this work.
Table of Contents
- 1. Introduction
- 2. Babase System Architecture
- 3. Baboon Data: Primary Source Material
- Group Membership and Life Events
- Physical Traits
- Sexual Cycles
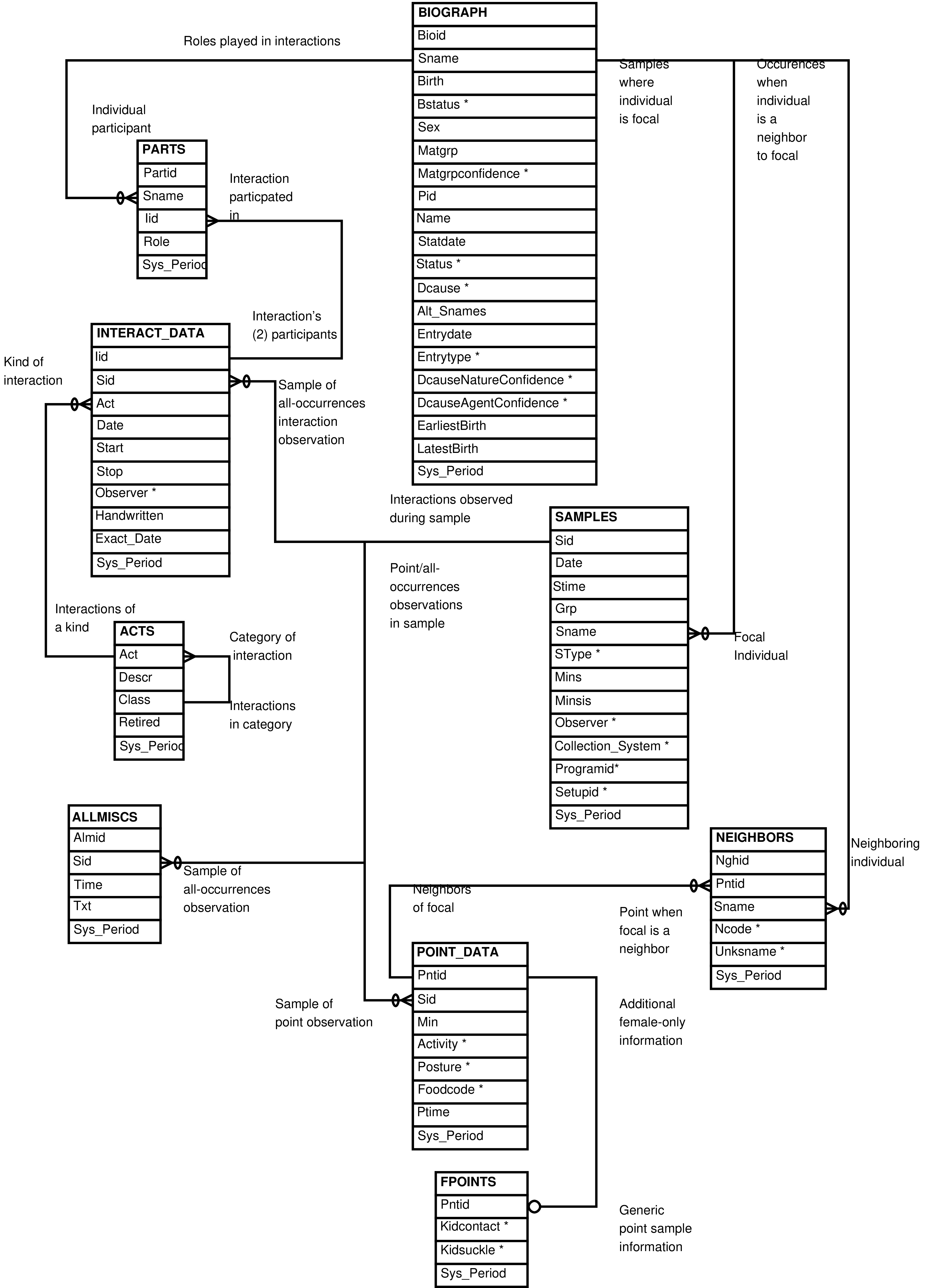
- Social and Multiparty Interactions
- ALLMISCS (Ad-libitum sample data)
- CONSORTS (multiparty disputes over CONSORTshipS)
- FPOINTS (Point data on Females)
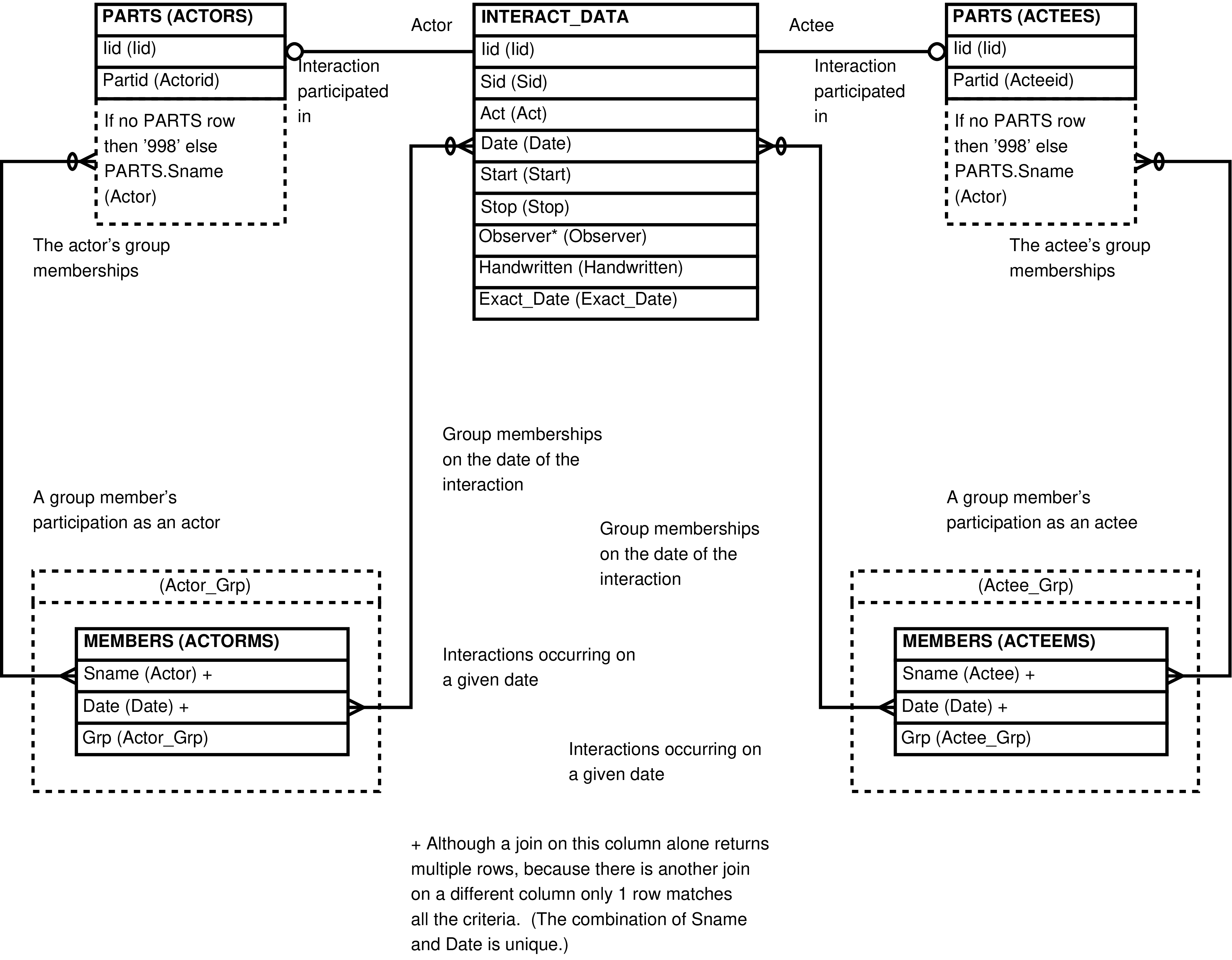
- INTERACT_DATA (Interactions)
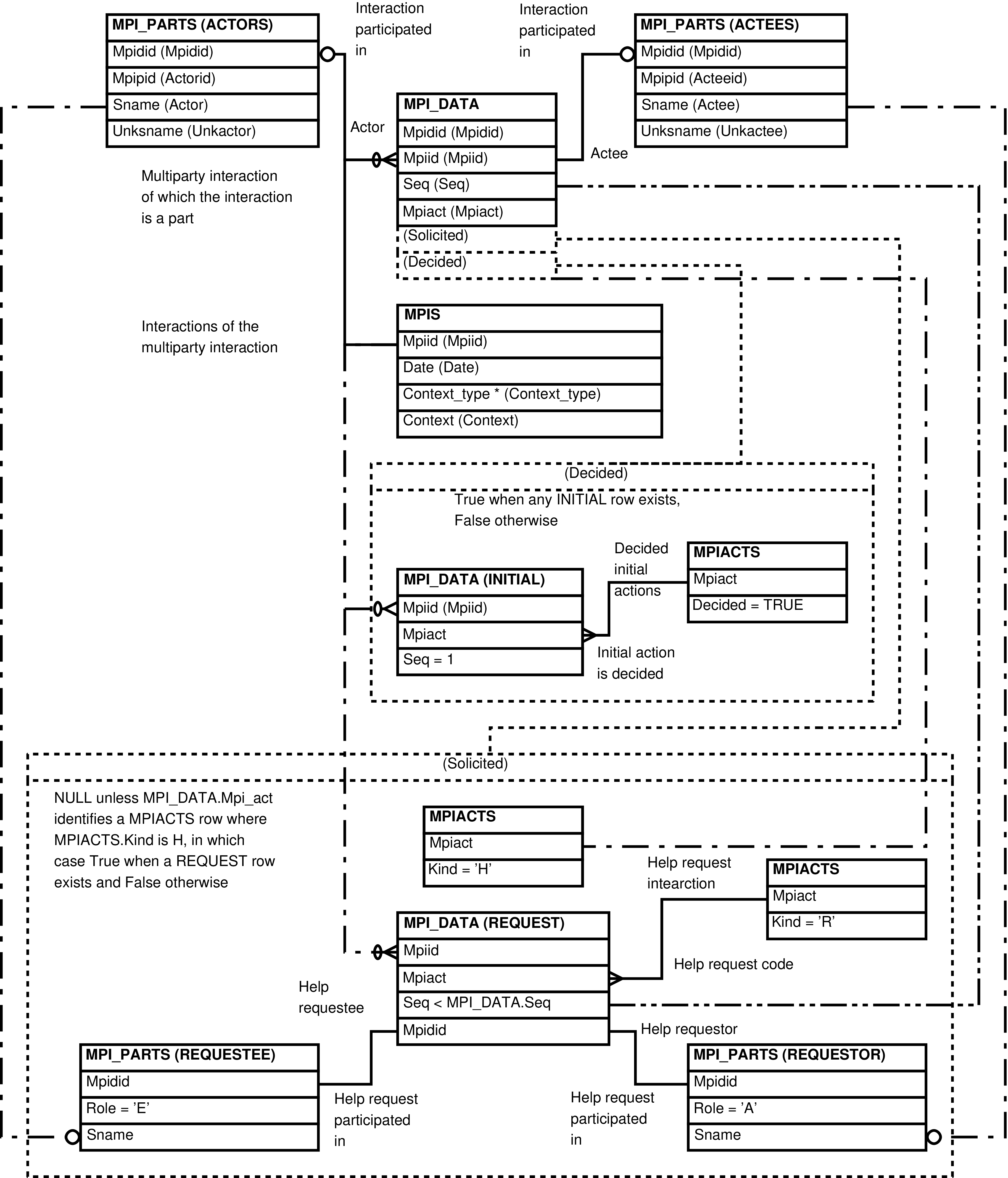
- MPIS (Multiparty InteractionS)
- MPI_DATA (Multiparty dyadic Interactions)
- MPI_PARTS (Multiparty Interaction PARTicipantS)
- PARTS (Participants in interactions)
- POINT_DATA (Point observation data)
- NEIGHBORS (point observation data on Neighbors)
- SAMPLES (all-occurrences Samples)
- Darting
- ANESTHS (Extra Sedation Administered During Darting)
- BODYTEMPS (Darting Body Temperature Measurements)
- CHESTS (Darting Chest Circumference Measurements)
- CROWNRUMPS (Darting Crown-to-Rump Measurements)
- DART_SAMPLES (Darting Tissue Sample Records)
- DARTINGS (Baboon Darting Events)
- DPHYS (Darting Physiological Measurements)
- HUMERUSES (Darting Humerus Length Measurements)
- PCVS (Darting Blood Measurements)
- TEETH (Darting Tooth Data)
- TESTES_ARC (Darting Testes circumference Data)
- TESTES_DIAM (Darting Testes Diameter Data)
- TICKS (Darting Tick and Parasite Data)
- ULNAS (Darting Ulna Length Measurements)
- VAGINAL_PHS (Darting Vaginal pH Measurements)
- Inventory
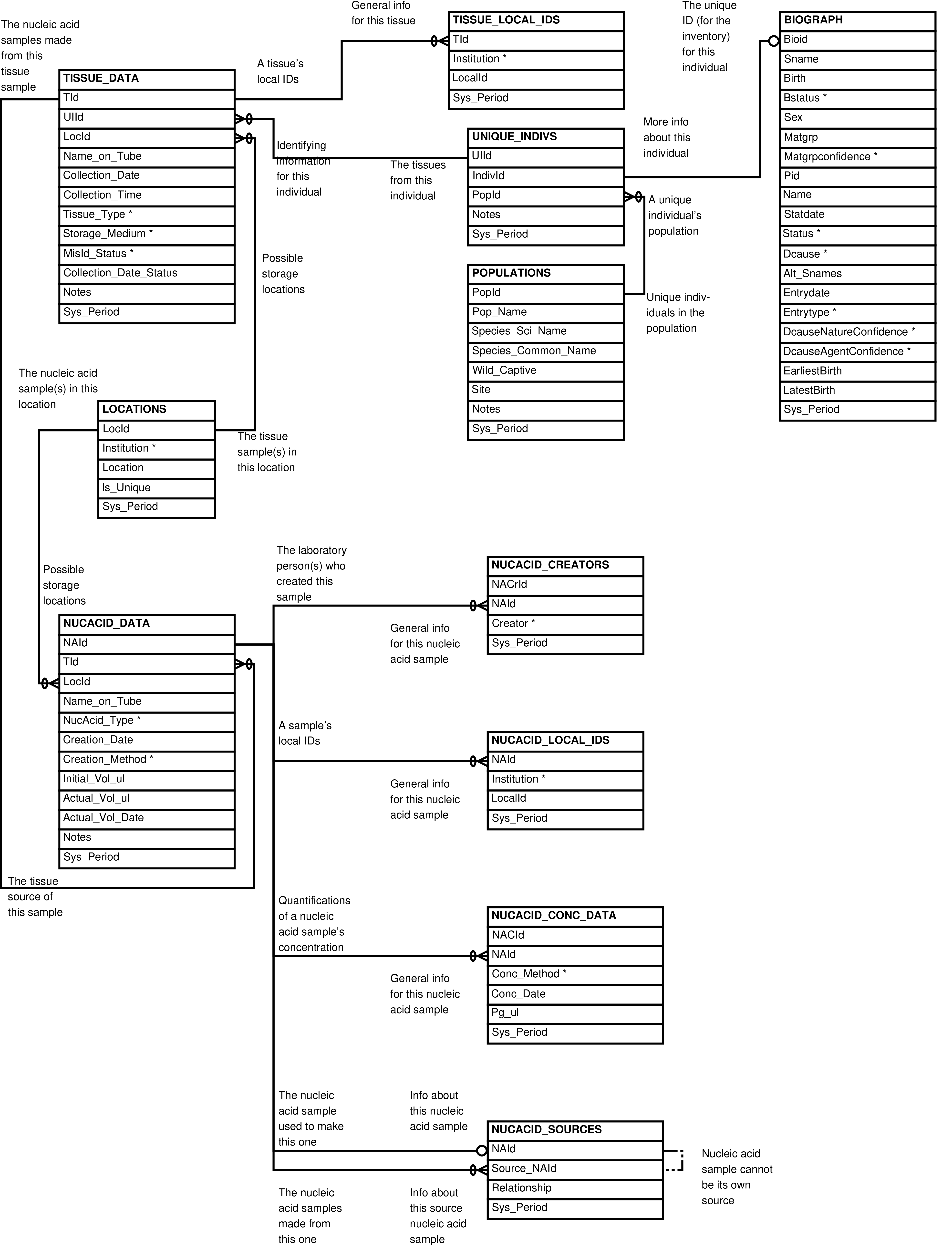
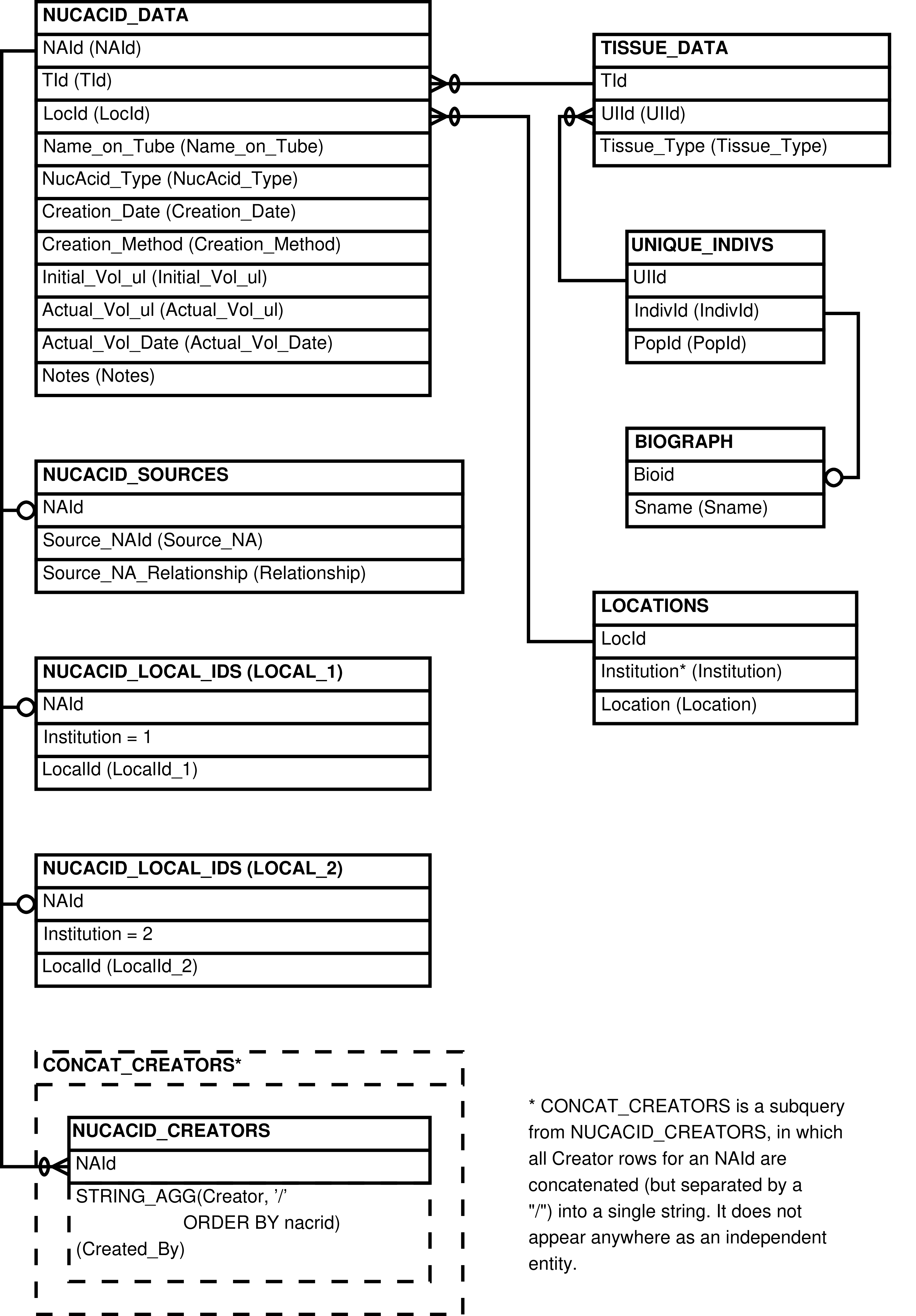
- LOCATIONS
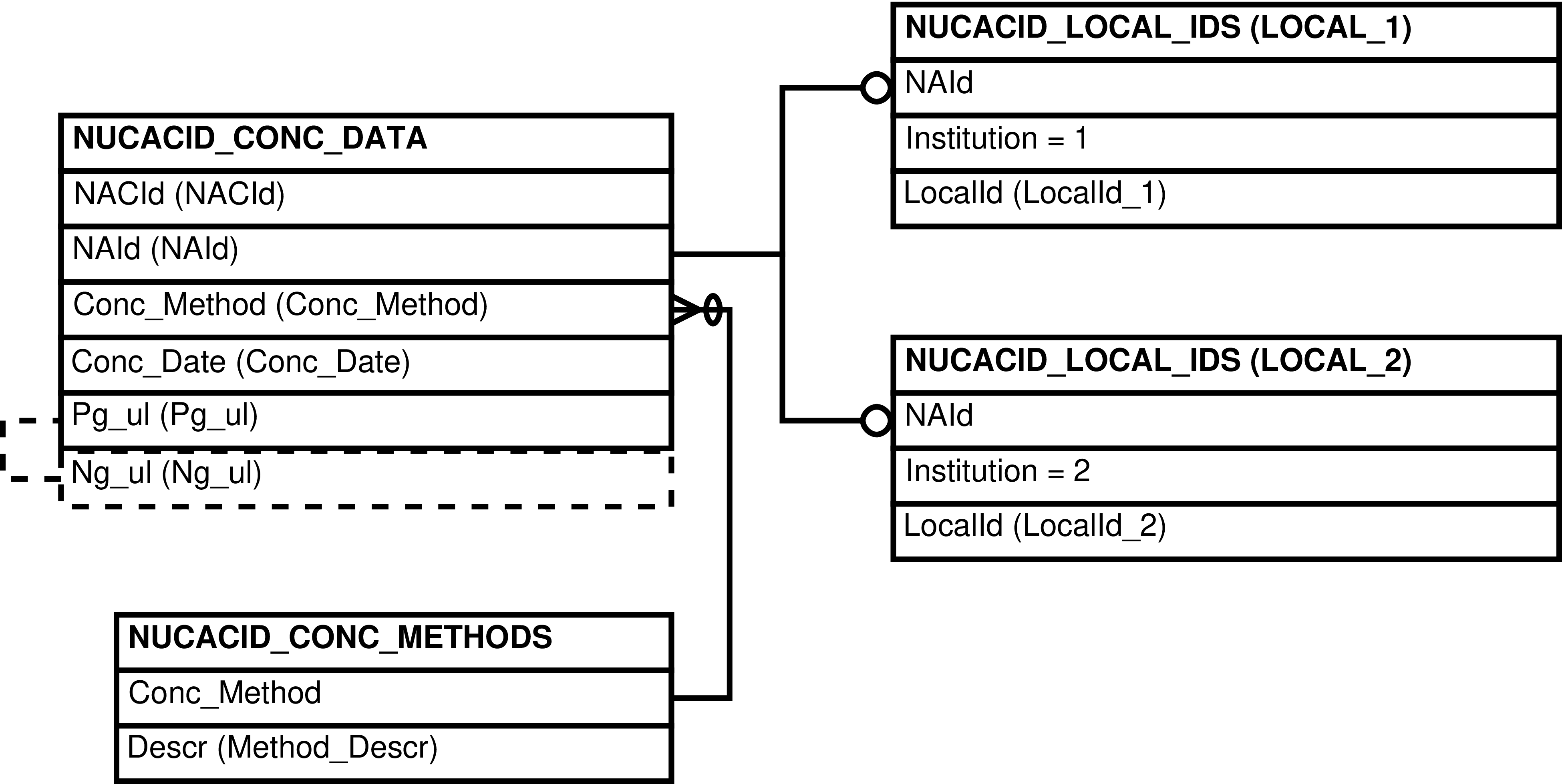
- NUCACID_CONC_DATA (NUCleic ACID CONCentration DATA)
- NUCACID_CREATORS (NUCleic ACID CREATORS)
- NUCACID_DATA (General information about NUCleic ACID samples)
- NUCACID_LOCAL_IDS (LOCAL IDentifierS for NUCleic ACID samples)
- NUCACID_SOURCES
- POPULATIONS
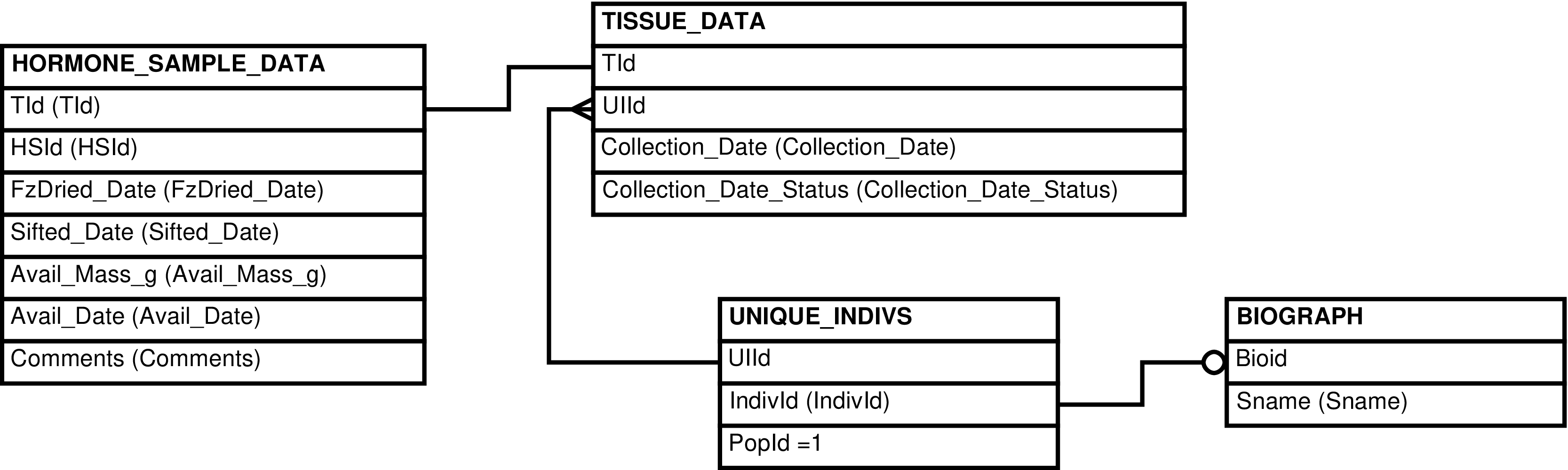
- TISSUE_DATA (General information about TISSUE samples)
- TISSUE_LOCAL_IDS (LOCAL IDentifierS for TISSUE samples)
- TISSUE_SOURCES
- UNIQUE_INDIVS (All UNIQUE INDIVidualS)
- SWERB Data (Group-level Geolocation Data)
- AERIALS (Aerial photos)
- GPS_UNITS (Individual GPS Devices)
- QUAD_DATA (map Quadrants)
- SWERB_BES (Begin/Ends: Uninterrupted bouts of group-level observation)
- SWERB_DATA (Group Level GPS Point Samples)
- SWERB_DEPARTS_DATA (Observation team departures from camp)
- SWERB_DEPARTS_GPS (SWERB GPS Departure data)
- SWERB_GWS (SWERB Grove and Waterholes)
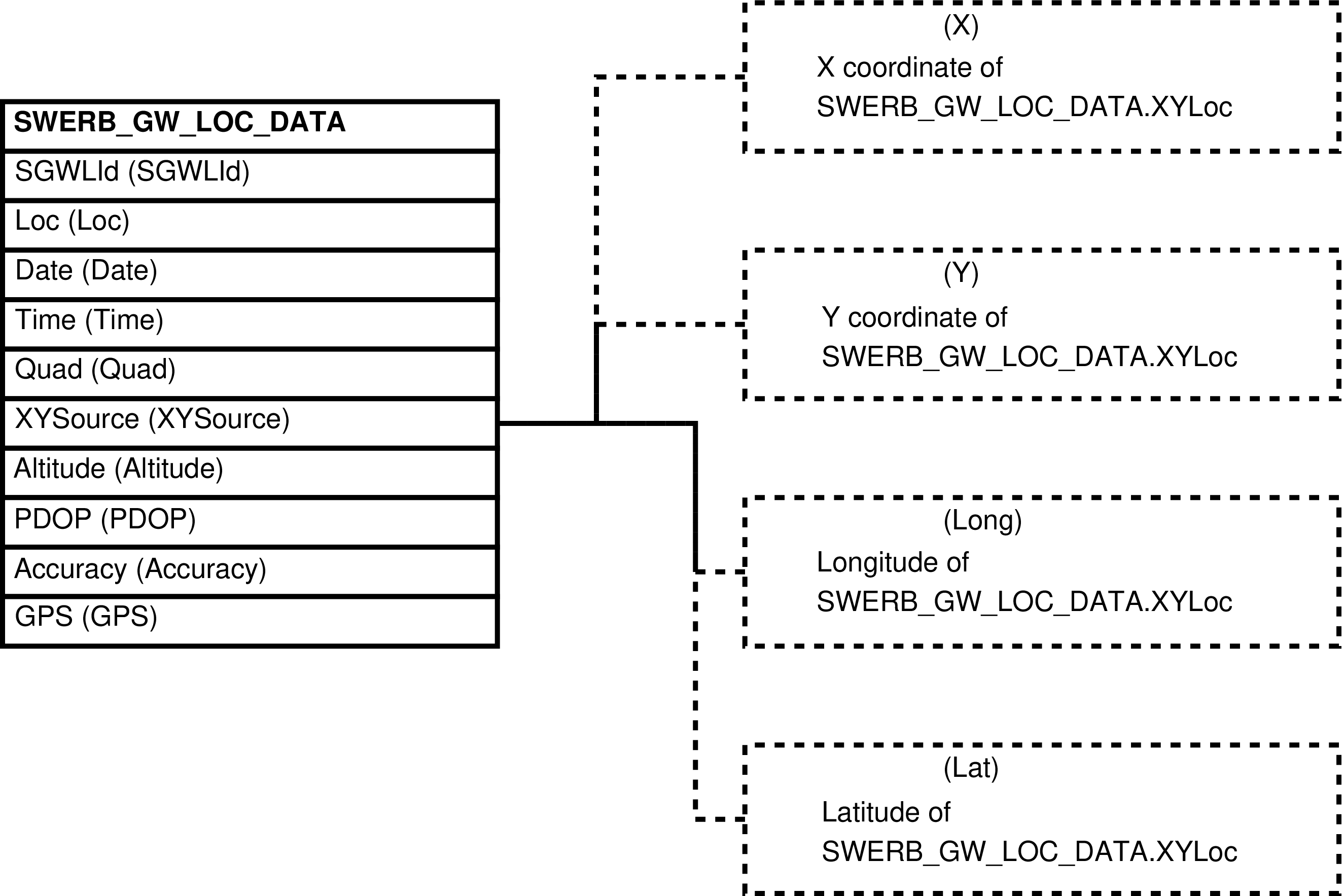
- SWERB_GW_LOC_DATA (SWERB Grove/Waterhole Location Data)
- SWERB_LOC_DATA (LOCation-Specific DATA from GPS Points)
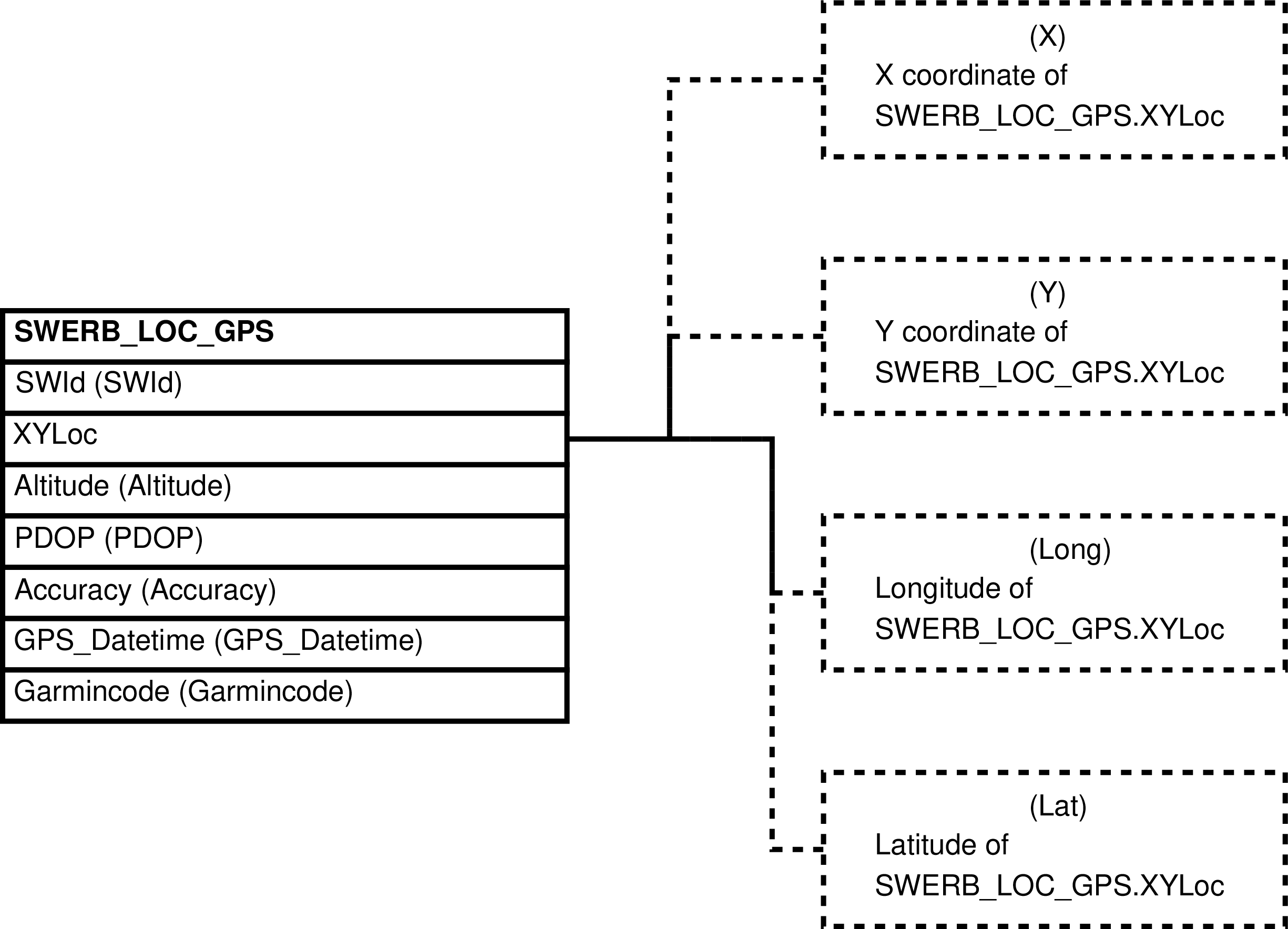
- SWERB_LOC_GPS (Secondary Data for LOCations in GPS points)
- SWERB_OBSERVERS
- TREES
- Weather Data
- Metadata
- 4. Baboon Data: Analyzed
- Darting
- Group Membership and Life Events
- Physical Traits
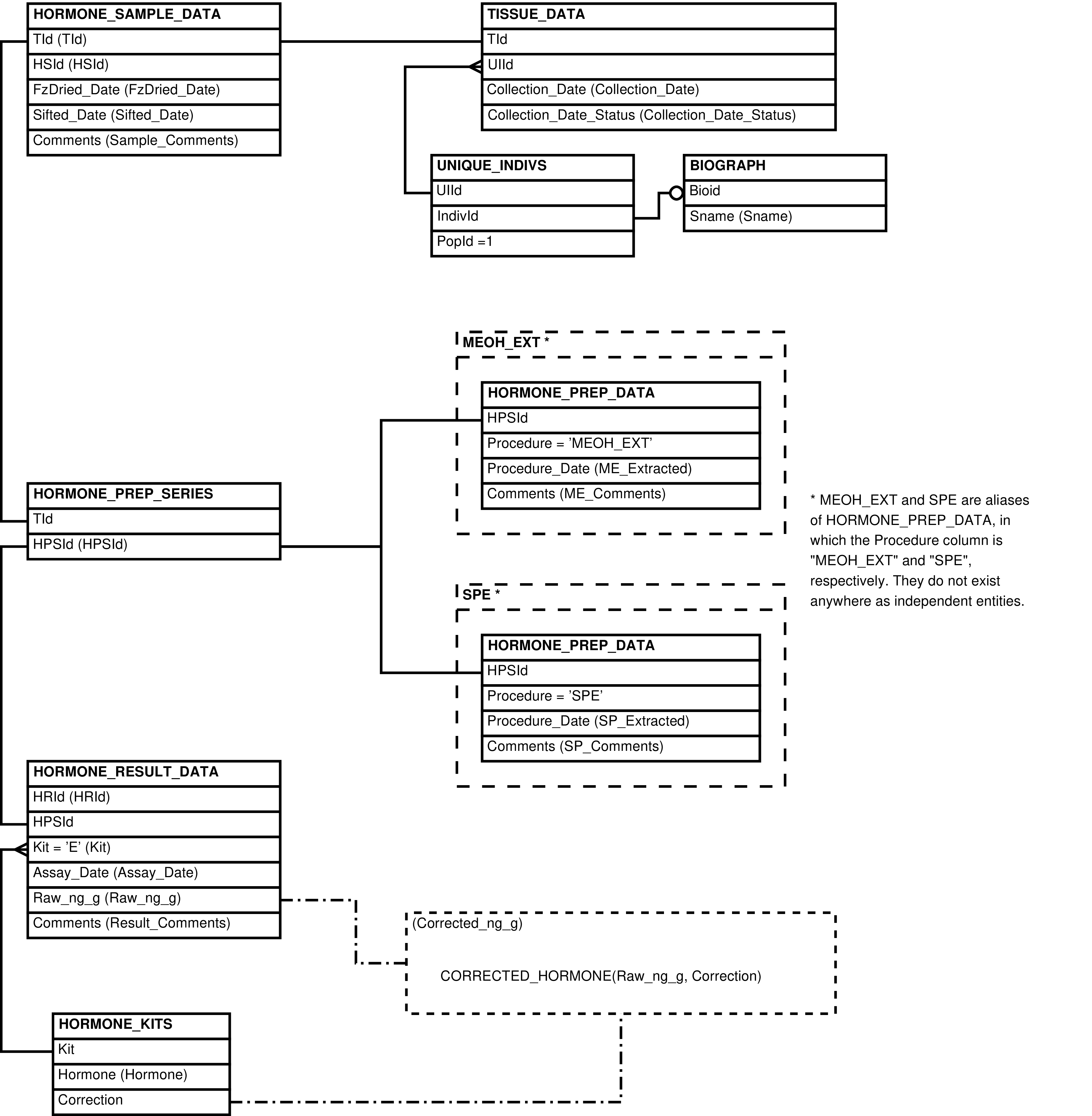
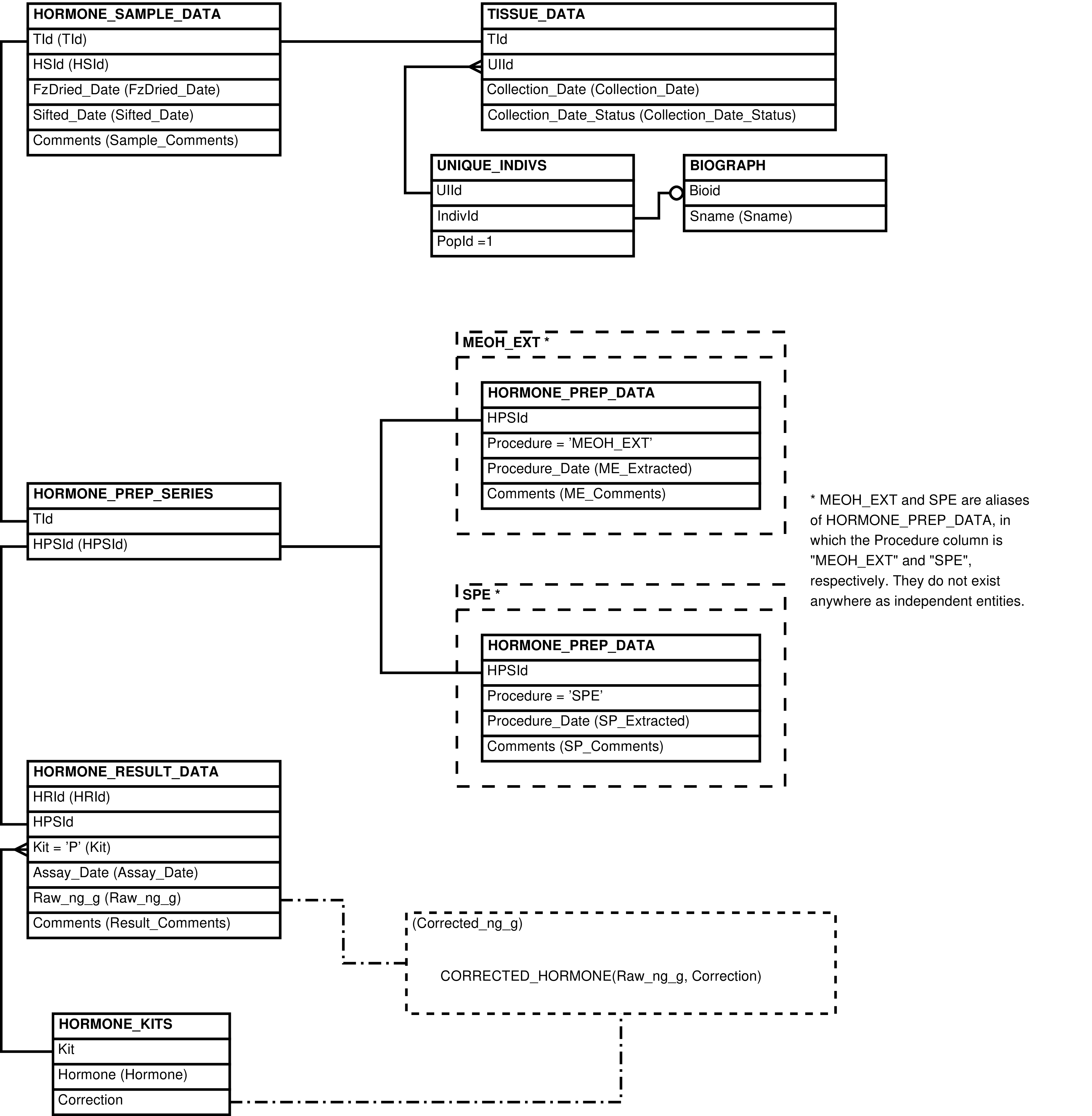
- HORMONE_KITS (KITS used to assay HORMONE concentration)
- HORMONE_PREP_DATA (Lab PREParations performed on hormone samples)
- HORMONE_PREP_SERIES (SERIES of Lab PREParations performed on hormone samples)
- HORMONE_RESULT_DATA (RESULTs of HORMONE concentration assays)
- HORMONE_SAMPLE_DATA (Tissue SAMPLEs used for HORMONE analysis)
- HYBRIDGENE_ANALYSES
- HYBRIDGENE_SCORES
- HYBRIDMORPH_OBSERVERS (Observers of hybrid morphology scores)
- HYBRIDMORPH_REPORTS (Hybrid morphology score collection events)
- HYBRIDMORPH_SCORE_DATA (Hybrid morphology scores)
- SWERB Data
- Interpolation
- The Social Group Residency Rules
- The Sexual Cycle Day-By-Day Tables
- Sexual Cycle Determination
- Automatic Sequencing
- Automatic Mdate Generation
- 5. Support Tables
- General Support Tables
- Group Membership and Life Events
- BSTATUSES (Birth Accuracy Indicators)
- CONFIDENCES (death cause (nature and agent), dispersal, and matgrp Confidence levels)
- DADS_COMPLETENESS (Completeness Scores in Paternity Assignments)
- DADS_EVIDENCE_TERMS (Types of data in paternity analyses)
- DADS_MISMATCHES (Types of Genetic Mismatches)
- DCAUSES (Causes of Death)
- DEMOG_REFERENCES (Demography Note References)
- DEATHNATURES (Natures of Death Causes)
- ENTRYTYPES (Categories of Entry to Study Population)
- MSTATUSES (Maturity Marker Statuses)
- RNKTYPES (Ranking Categories)
- STATUSES (Indicators of Record and Baboon Vividity)
- Physical Traits
- Social And Multiparty Interactions
- ACTIVITIES
- ACTS (Interaction Types)
- DATA_STRUCTURES (Data structures produced by Psion devices)
- CONTEXT_TYPES (multiparty Interaction Context Categories)
- FOODCODES (Food item Codes)
- FOODTYPES (Food Types)
- KIDCONTACTS (spatial relationship between mother and infant)
- MPIACTS (Multiparty Interaction Types)
- NCODES (Neighbor classifications)
- PARTUNKS (problem identifying a multiparty interaction participant)
- POSTURES
- PROGRAMIDS (Program used on the device)
- SAMPLES_COLLECTION_SYSTEMS
- SETUPIDS (Setup files used in a data collection program)
- STYPES (Focal Sample Types)
- STYPES_ACTIVITIES (Activity values that are used with each SType)
- STYPES_NCODES (Ncodes that are used with each SType)
- STYPES_POSTURES (Postures that are used with each SType)
- SUCKLES (infant suckling activity)
- Sexual Cycles and The Sexual Cycle Day-By-Day Tables
- Darting
- DART_SAMPLE_CATS (Darting Sample Categories)
- DART_SAMPLE_TYPES (Sample Types)
- DRUGS (darting anesthetics)
- LYMPHSTATES (Lymph node conditions)
- PARASITES (Parasites and their indicators)
- TCONDITIONS (Tooth Conditions)
- TICKSTATUSES (parasite count classifications)
- TOOTHCODES (kinds of teeth)
- TOOTHSITES (Locations of deciduous or adult teeth)
- TSTATES (State of Tooth existence)
- Inventory
- INSTITUTIONS
- MISID_STATUSES (MISIDentification STATUSES)
- NUCACID_CONC_METHODS (NUCleic ACID CONCentration quantification METHODS)
- NUCACID_CONC_UNITS (NUCleic ACID CONCentration quantification UNITS)
- NUCACID_CREATION_METHODS (NUCleic ACID CREATION METHODS)
- NUCACID_TYPES (NUCleic ACID TYPES)
- STORAGE_MEDIA
- TISSUE_TYPES
- SWERB Data
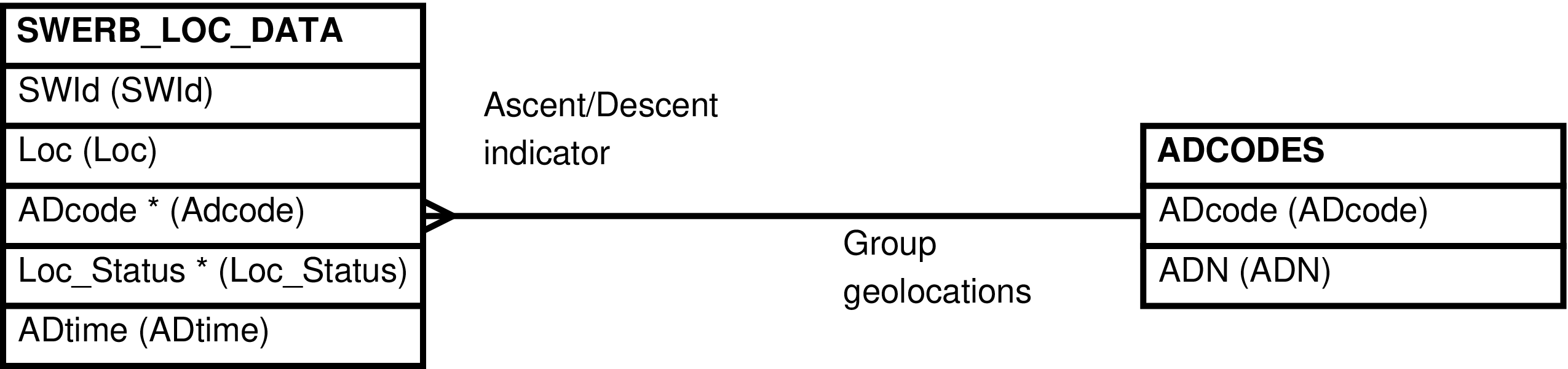
- ADCODES (SWERB Ascent and Descent relationships)
- PLACE_TYPES (codes for various landscape features)
- PREDATORS (codes for observed predators)
- SWERB_LOC_CONFIDENCES (SWERB Location Confidence Values)
- SWERB_LOC_STATUSES (SWERB Location Statuses)
- SWERB_TIME_SOURCES (SWERB Time Sources)
- SWERB_XYSOURCES (SWERB Time Sources)
- Weather Data
- Metadata
- 6. The Babase Views
- Group Membership and Life Events
- CENSUS_DEMOG (CENSUS extended with DEMOG information)
- CENSUS_DEMOG_SORTED (CENSUS_DEMOG, Sorted)
- CYCPOINTS_CYCLES (CYCPOINTS extended with CYCLES information)
- CYCPOINTS_CYCLES_SORTED (CYCPOINTS_CYCLES, Sorted)
- DEMOG_CENSUS (DEMOG, showing CENSUS information)
- DEMOG_CENSUS_SORTED (DEMOG_CENSUS, Sorted)
- GROUPS_HISTORY
- PARENTS
- POTENTIAL_DADS (Potential Dads)
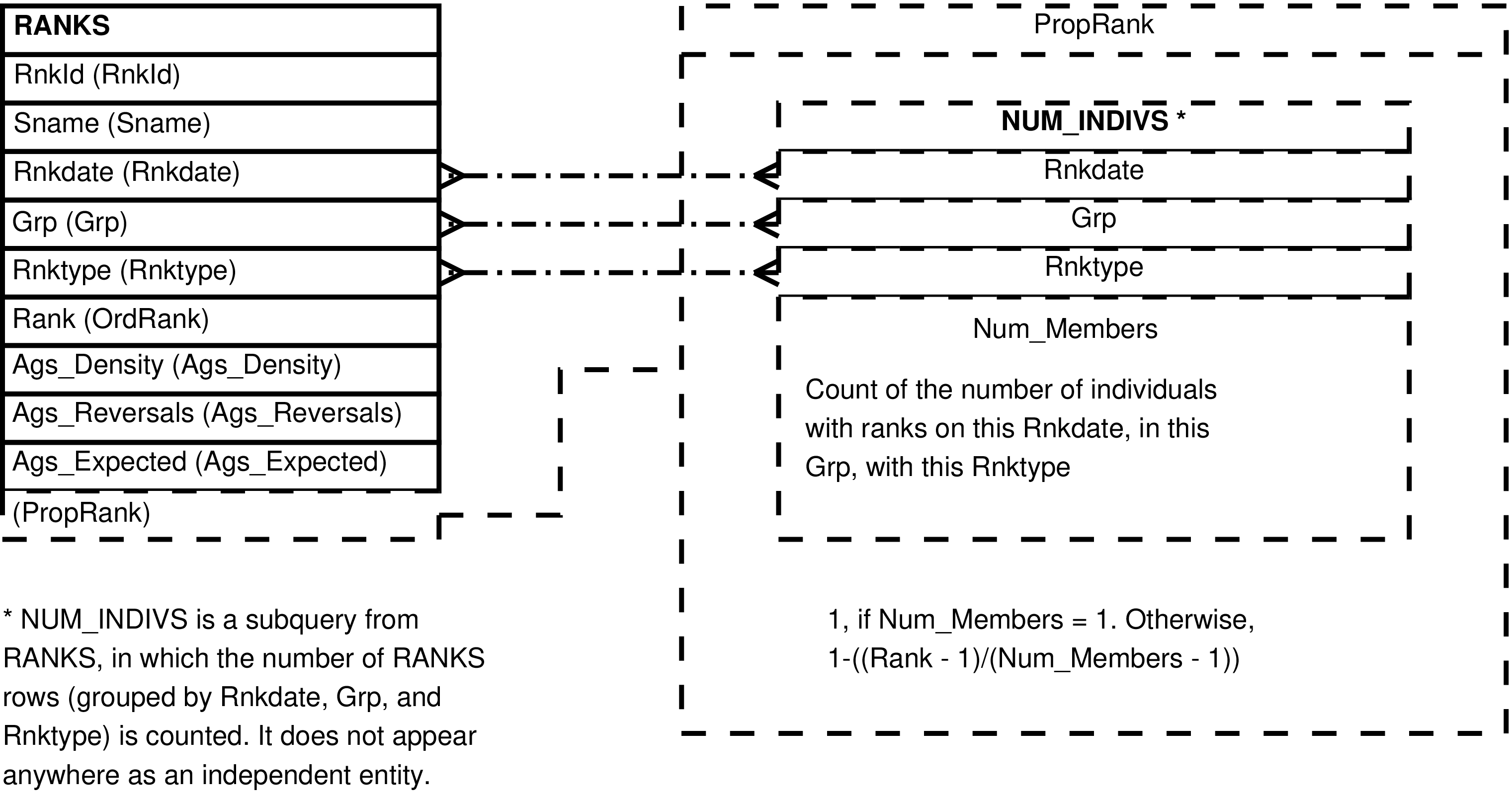
- PROPORTIONAL_RANKS (RANKS extended with calculated PROPORTIONAL ranks)
- Physical Traits
- ESTROGENS
- GLUCOCORTICOIDS
- HORMONE_PREPS
- HORMONE_RESULTS
- HORMONE_SAMPLES
- HYBRIDMORPH_SCORES
- PROGESTERONES
- TESTOSTERONES
- THYROID_HORMONES
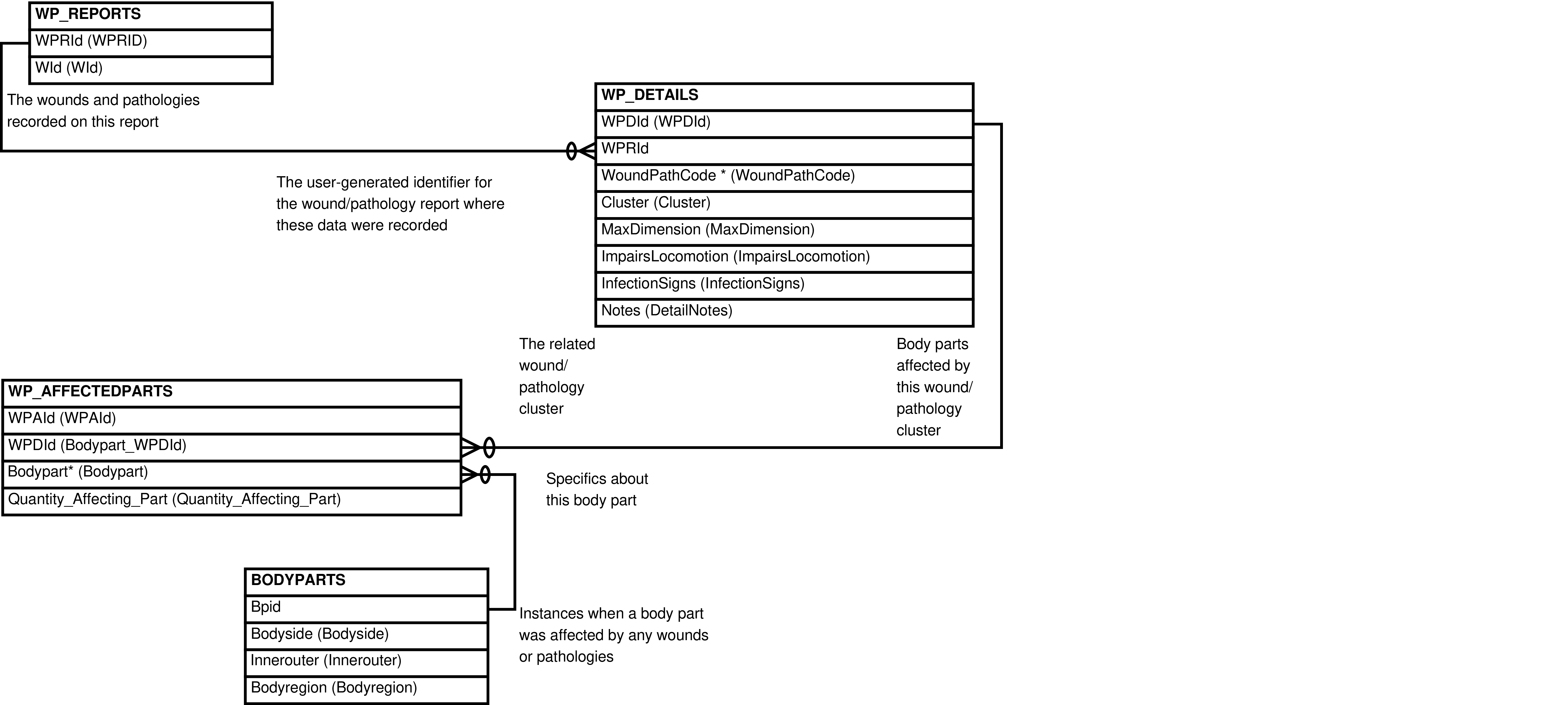
- WOUNDSPATHOLOGIES (All Wound/Pathology Data, Together)
- WP_DETAILS_AFFECTEDPARTS (WP_DETAILS, extended with WP_AFFECTEDPARTS)
- WP_HEALS (WP_HEALUPDATES, extended)
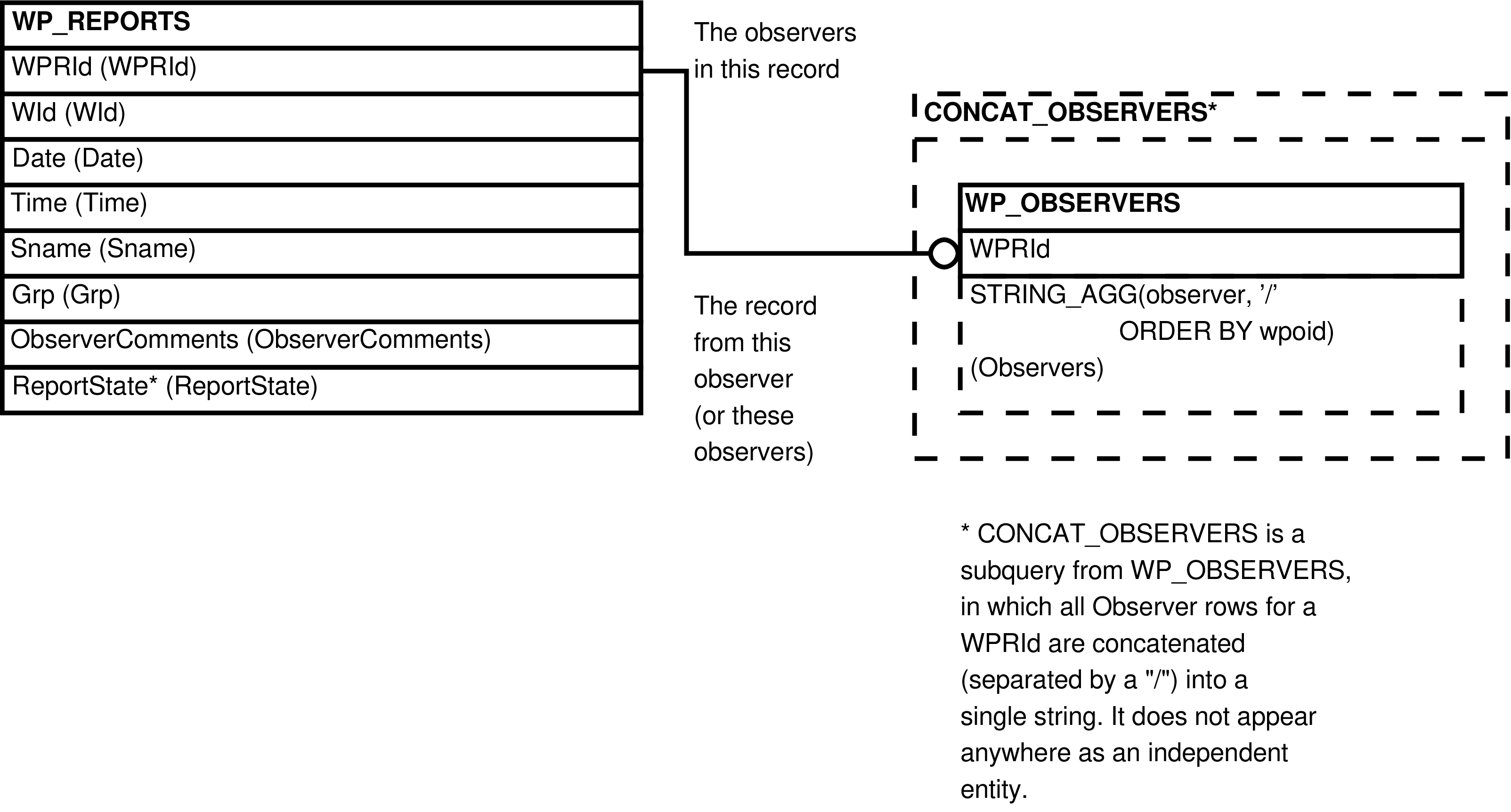
- WP_REPORTS_OBSERVERS (WP_REPORTS, extended with WP_OBSERVERS)
- Sexual Cycles
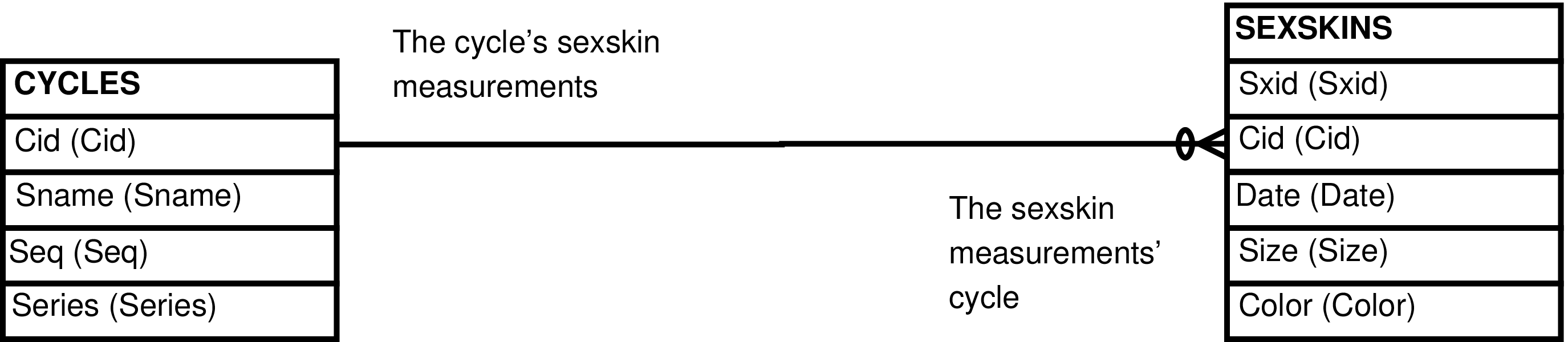
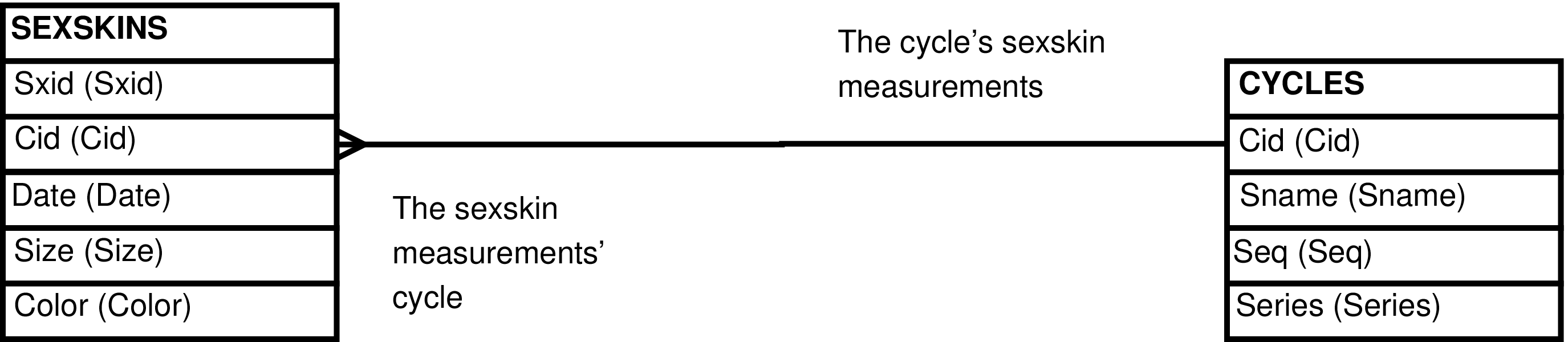
- CYCLES_SEXSKINS (CYCLES extended with SEXSKINS information)
- CYCLES_SEXSKINS_SORTED (CYCLES_SEXSKINS, Sorted)
- MATERNITIES (completed reproductive events)
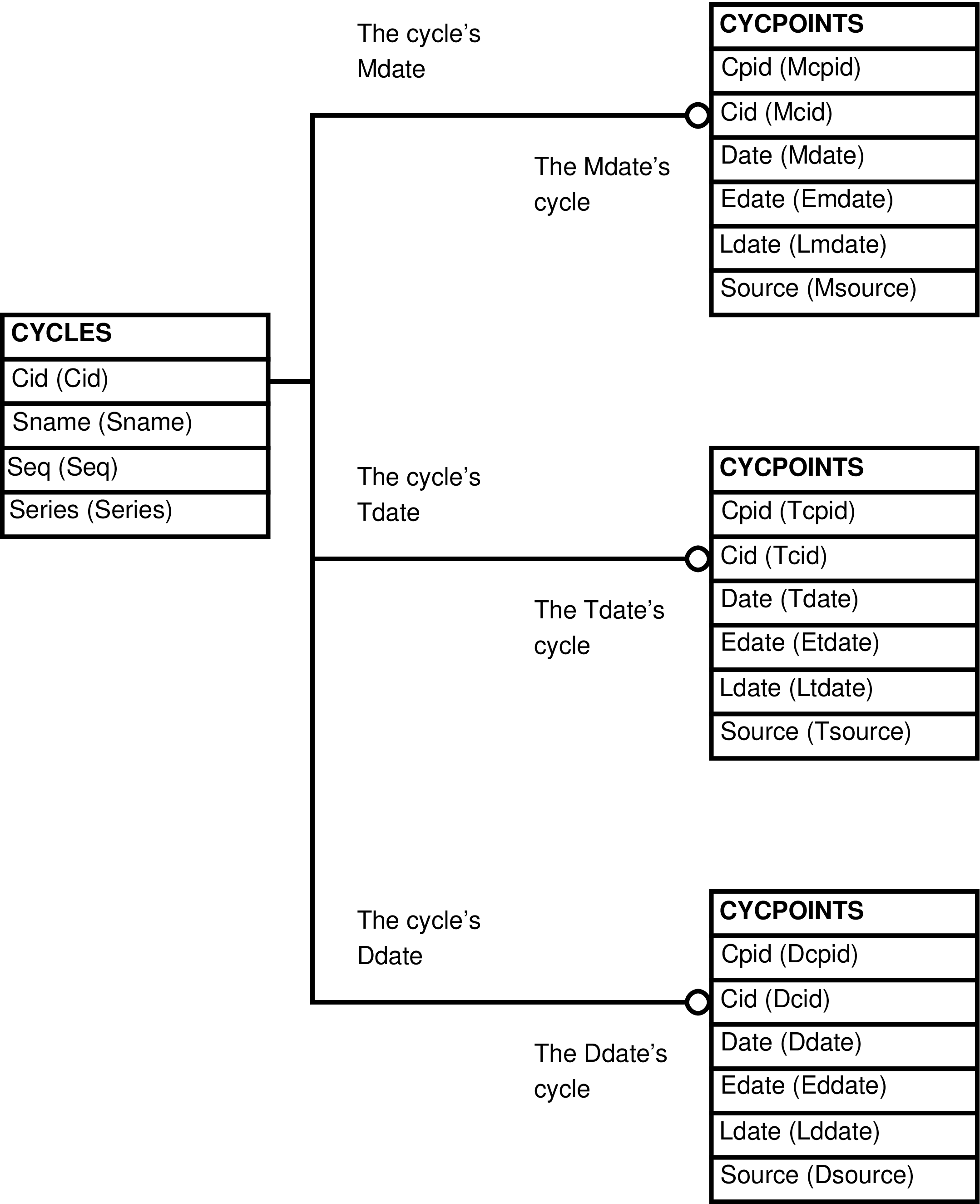
- MTD_CYCLES (CYCLES and Mdate, Tdate, and Ddate CYCPOINTS data)
- SEXSKINS_CYCLES (CYCLES extended with SEXSKINS information)
- SEXSKINS_CYCLES_SORTED (SEXSKINS_CYCLES, Sorted)
- SEXSKINS_REPRO_NOTES (SEXSKINS extended with REPRO_NOTES)
- Social and Multiparty Interactions
- ACTOR_ACTEES (Complete social interactions, INTERACT_DATA extended twice with PARTS)
- INTERACT (INTERACT_DATA, with enhanced dates and times)
- INTERACT_SORTED
- MPI_EVENTS (Dyadic social interactions that comprise multiparty interaction collections, MPIS joined with MPI_DATA extended twice with MPI_PARTS)
- MPI_UPLOAD: Upload Multiparty Interactions
- POINTS (POINT_DATA, with enhanced times)
- POINTS_SORTED (POINTS, Sorted)
- SAMPLES_GOFF (SAMPLES, with the Group OF the Focal)
- Darting
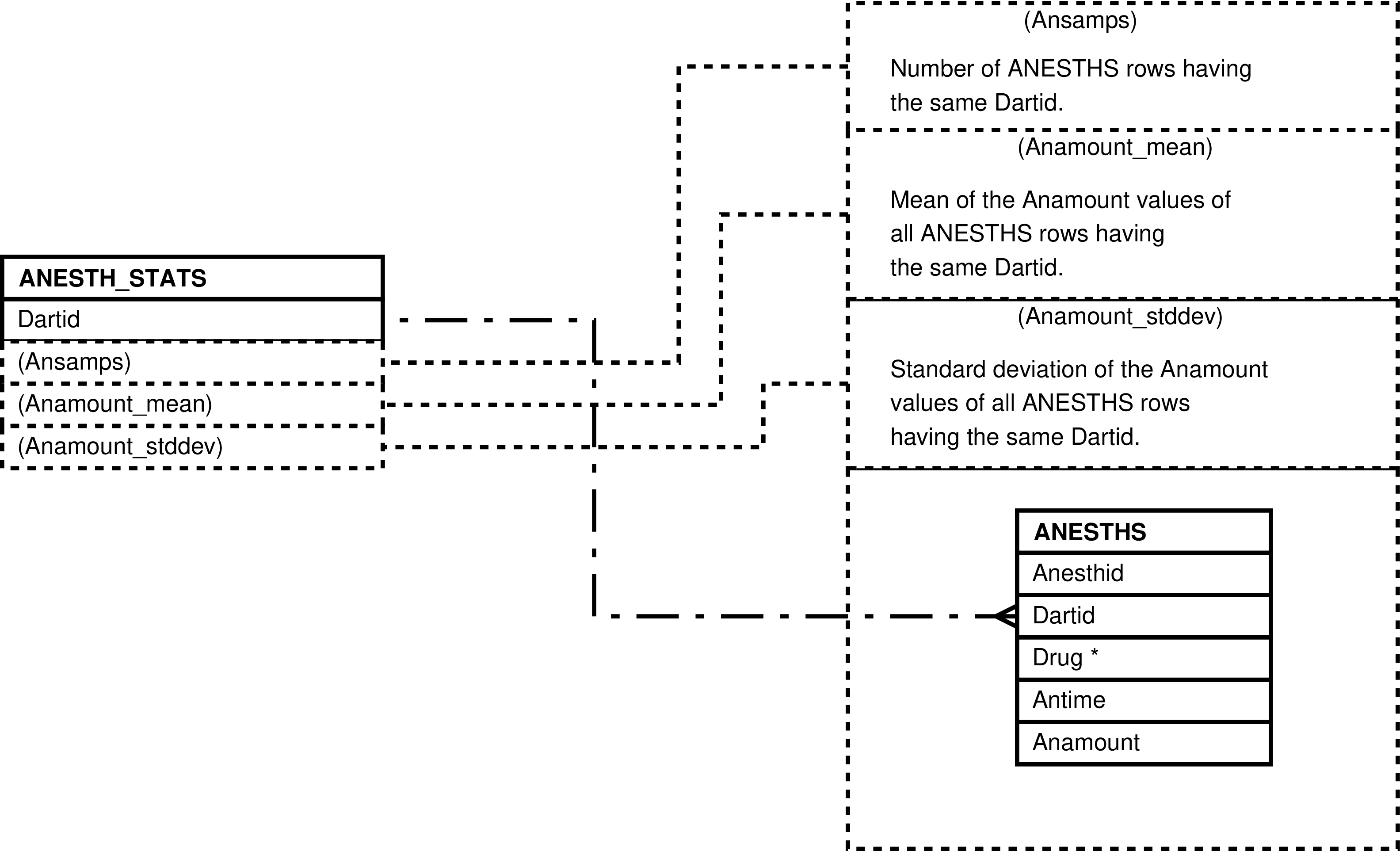
- ANESTH_STATS (darting additional Anesthetic Statistics)
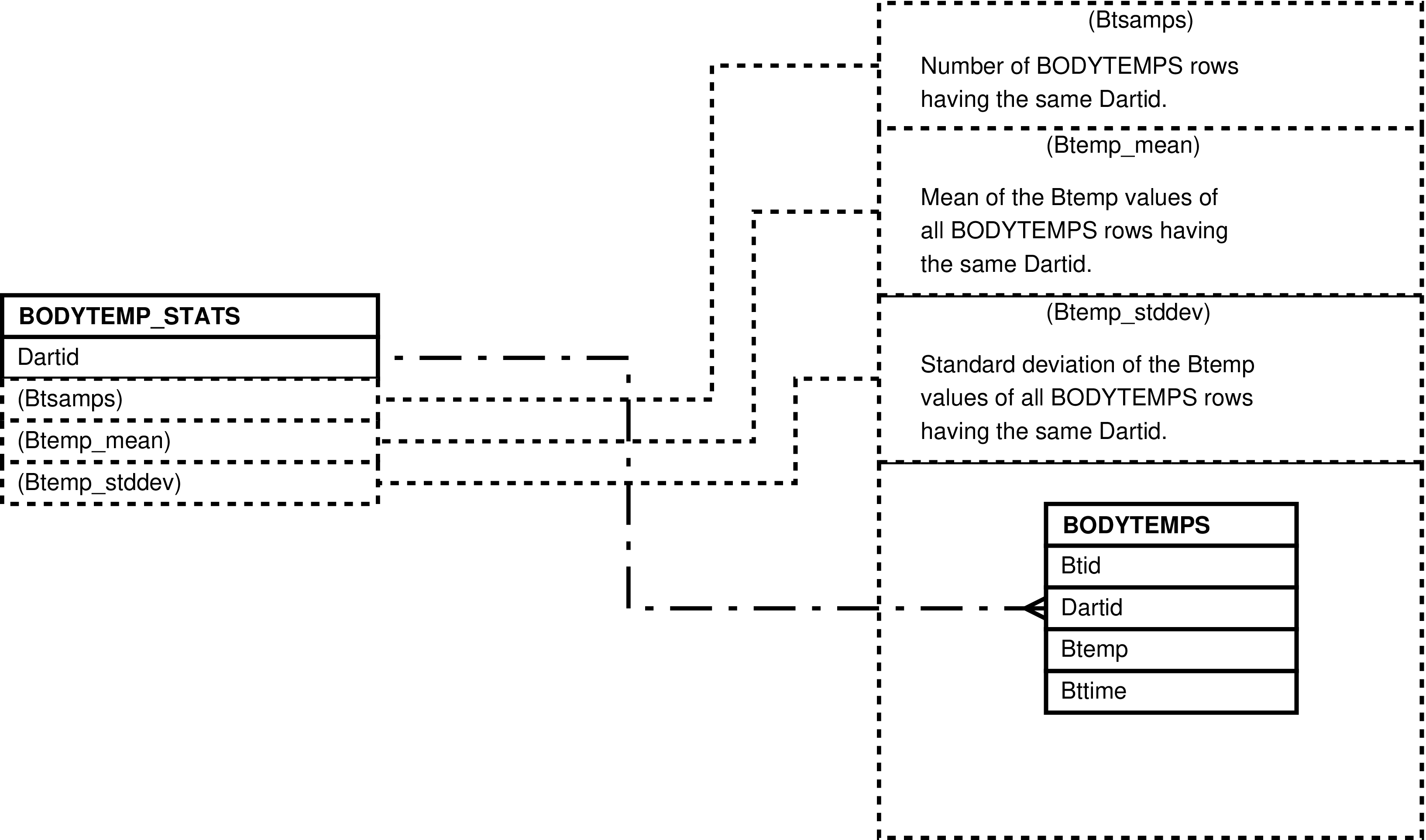
- BODYTEMP_STATS (darting Body Temperature Statistics)
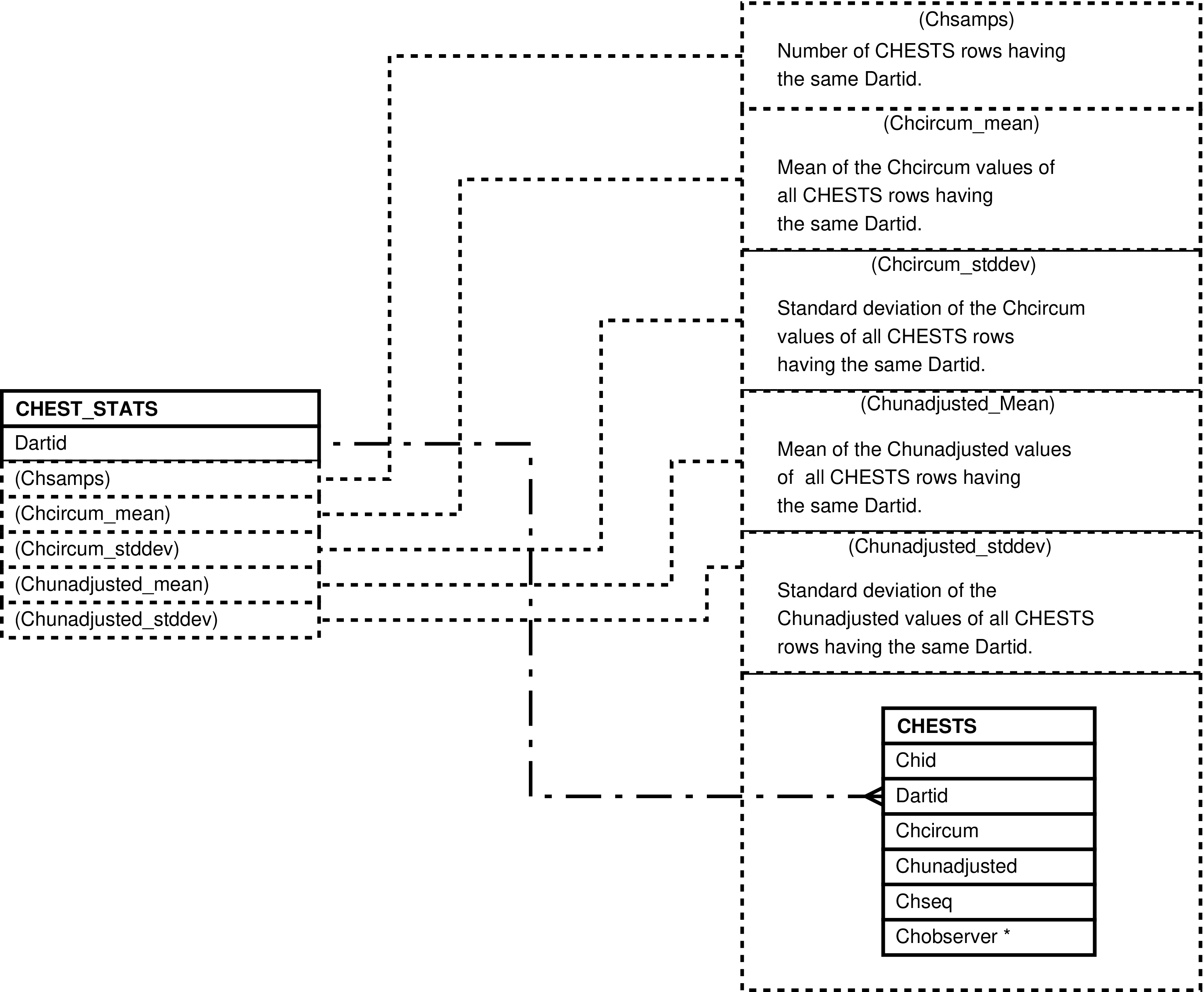
- CHEST_STATS (darting Chest circumference Statistics)
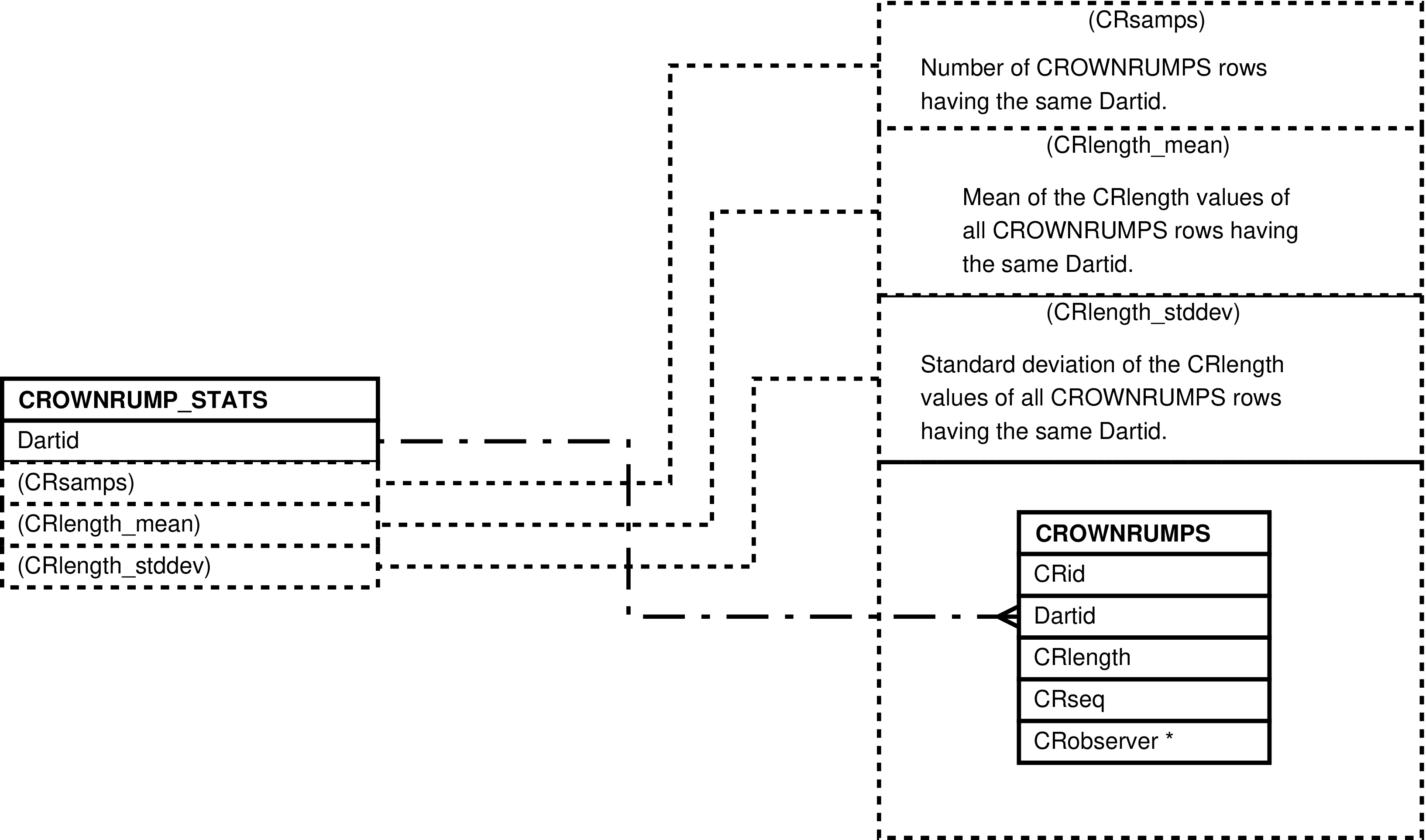
- CROWNRUMP_STATS (darting Crown-to-Rump Statistics)
- DART_FLOW_CYTOMETRY_UPLOAD (facility for recording flow cytometry data)
- DART_LOGISTICS_UPLOAD (facility for adding data about the logistics of a darting)
- DART_MORPHOLOGY_UPLOAD (facility for adding morphology data collected during a darting)
- DART_PHYSIOLOGY_UPLOAD (facility for adding physiology data collected during a darting)
- DART_SAMPLES_UPLOAD (facility for recording tissue samples collected during a darting)
- DART_TEETH_UPLOAD (facility for adding tooth data collected during a darting)
- DART_TESTES_ARC_UPLOAD (facility for adding testicle circumference data)
- DART_TESTES_DIAM_UPLOAD (facility for adding testicle diameter data)
- DART_TICKS_UPLOAD (facility for adding data from parasite counts performed during a darting)
- DART_VAGINAL_PHS_UPLOAD (facility for recording vaginal pH measurements performed during a darting)
- DART_WBC_COUNTS_UPLOAD (facility for recording white blood cell counts from blood smears)
- DSAMPLES (darting sample records with columns for each sample type)
- DENT_CODES (darting Dentition records with columns for each Toothcode)
- DENT_SITES (darting Dentition records with columns for each Toothsite)
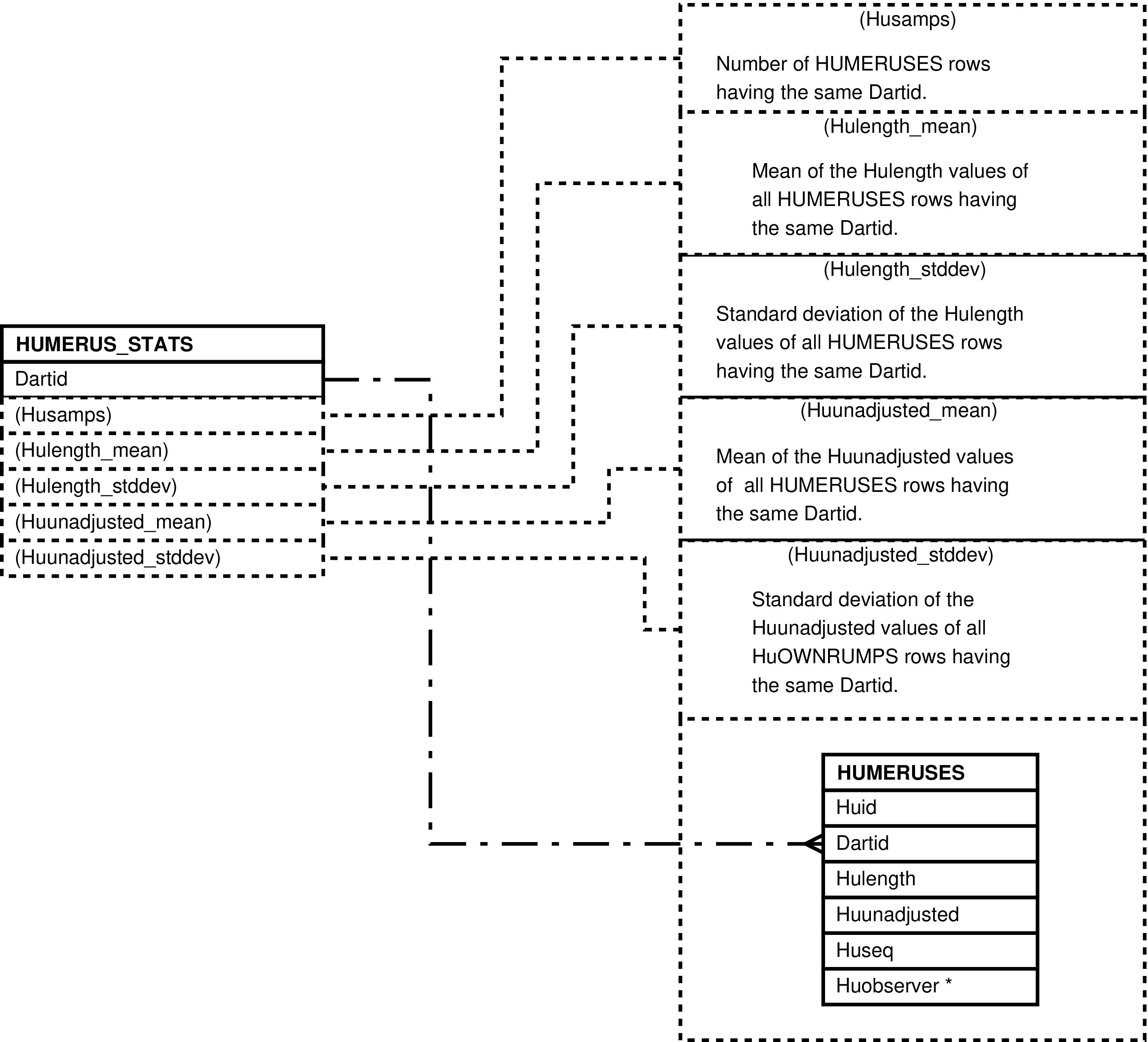
- HUMERUS_STATS (darting Humerus length Statistics)
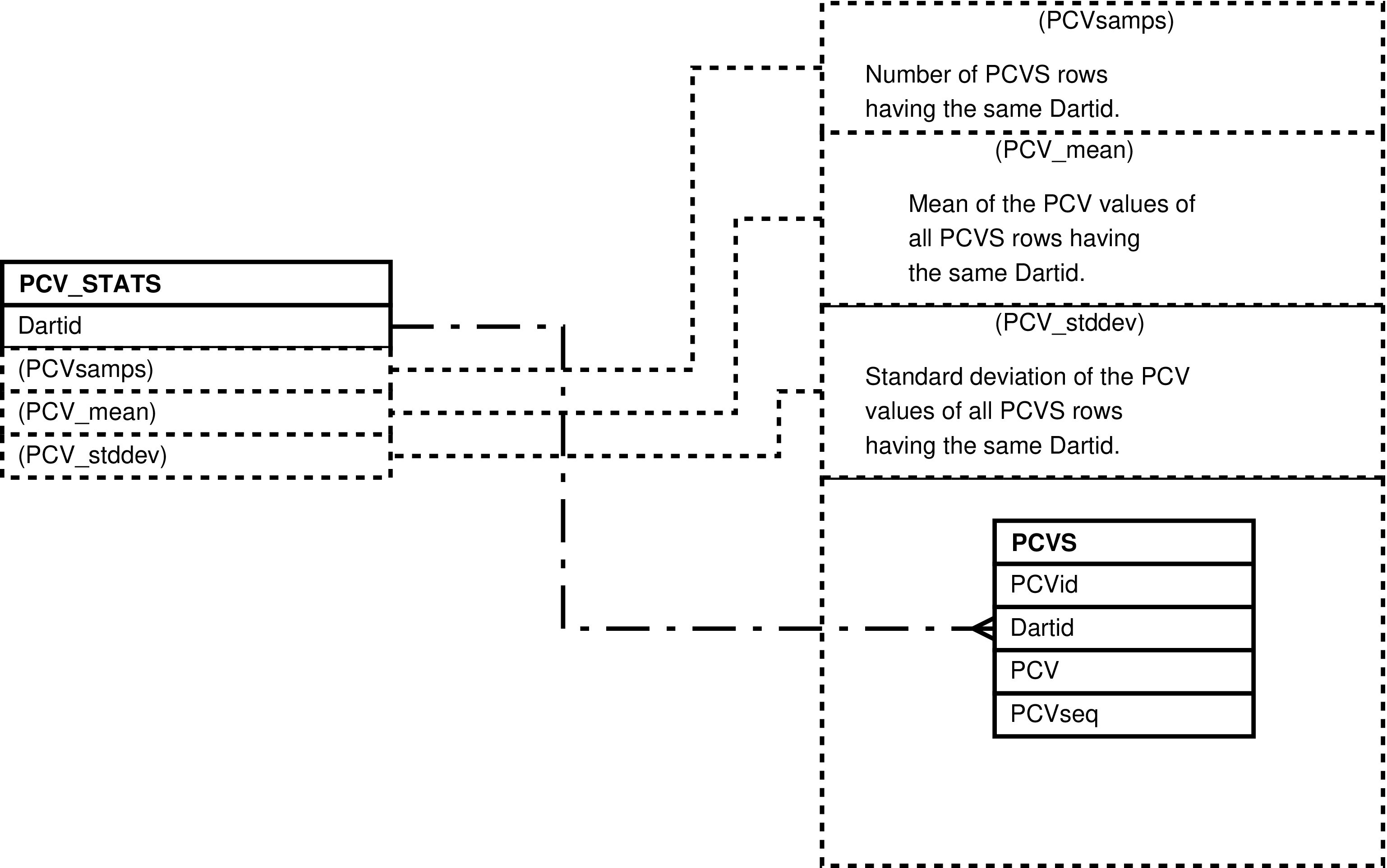
- PCV_STATS (darting PCV Statistics)
- TESTES_ARC_STATS (darting Testes circumference Statistics)
- TESTES_DIAM_STATS (darting Testes Diameter Statistics)
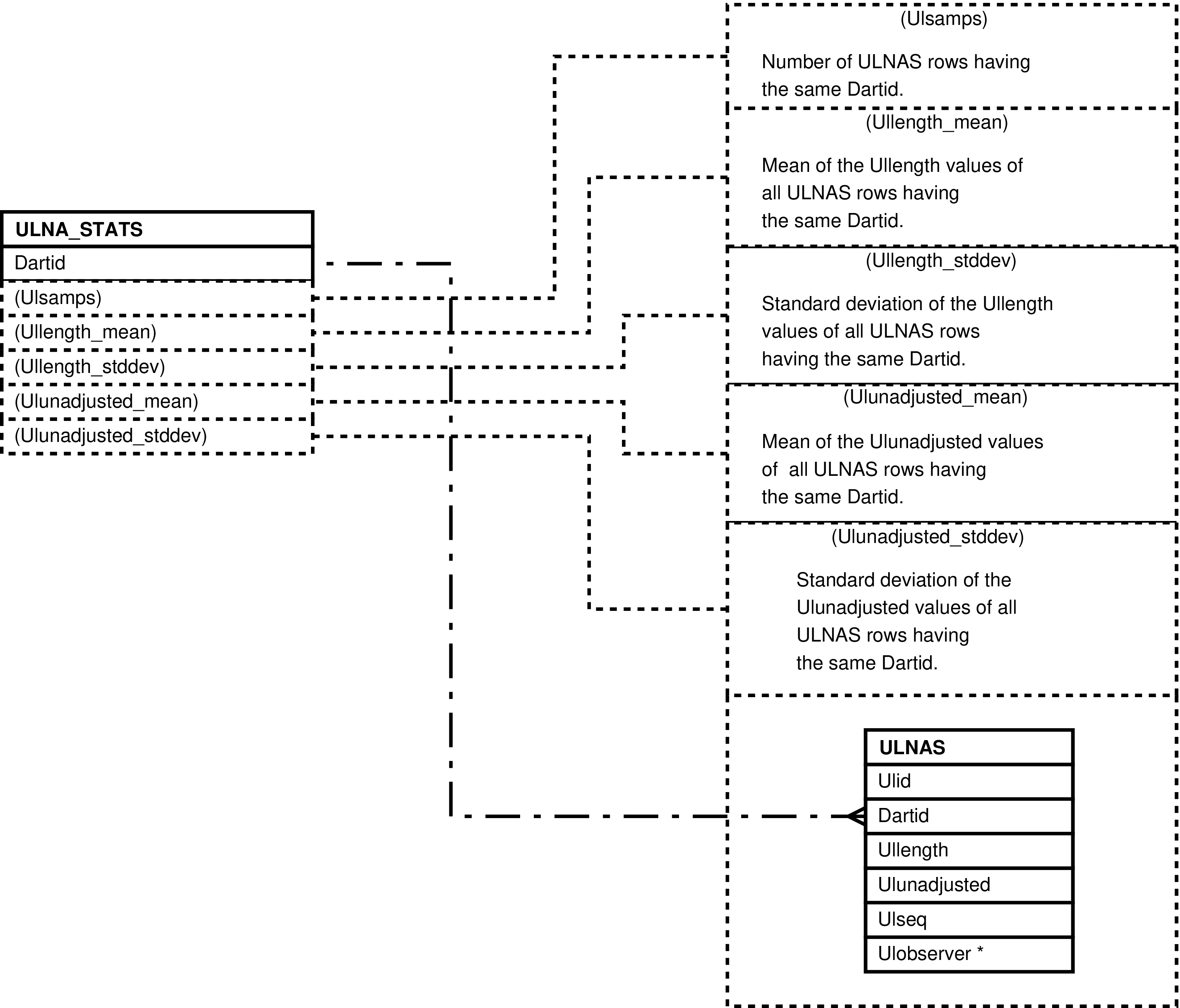
- ULNA_STATS (darting Ulna length Statistics)
- VAGINAL_PH_STATS (darting Vaginal pH Statistics)
- Inventory
- SWERB Data (Group-level Geolocation Data)
- QUADS (map Quadrants)
- SWERB (Group level gps point samples)
- SWERB_DATA_XY (The SWERB_DATA table with separate X and Y coordinates)
- SWERB_DEPARTS (SWERB observation team Departures from camp)
- SWERB_GW_LOCS (SWERB Grove and Waterhole Locations)
- SWERB_GW_LOC_DATA_XY (The SWERB_GW_LOC_DATA table with separate X and Y coordinates)
- SWERB_LOC_GPS_XY (The SWERB_LOC_GPS table with separate X and Y coordinates)
- SWERB_LOCS (placement of a group at a landscape feature)
- SWERB_UPLOAD (facility for uploading data into SWERB)
- Weather Data
- Views Which Add Gid To Tables
- The BIRTH_GRP View
- The ENTRYDATE_GRP View
- The STATDATE_GRP View
- The CONSORTDATES_GRP View
- The CYCGAPDAYS_GRP View
- The CYCGAPS_GRP View
- The CYCSTATS_GRP View
- The DARTINGS_GRP View
- The DISPERSEDATES_GRP View
- The MATUREDATES_GRP View
- The MDINTERVALS_GRP View
- The MMINTERVALS_GRP View
- The RANKDATES_GRP View
- The REPSTATS_GRP View
- 7. Data Entry
- 8. Babase Programs
- A. Manipulating Date and Time Values
- B. Querying-All-Occurrences-Interactions
- C. Alteration Of Sexual Cycle Ids (Cid)
- D. Babase Revision History
- E. DocBook, Styling and other issues
- F. Restrictions: Things Not To Do
- G. Database Transactions Explained
- H. The Warning System
- I. Temporal Tables and babase_history
List of Figures
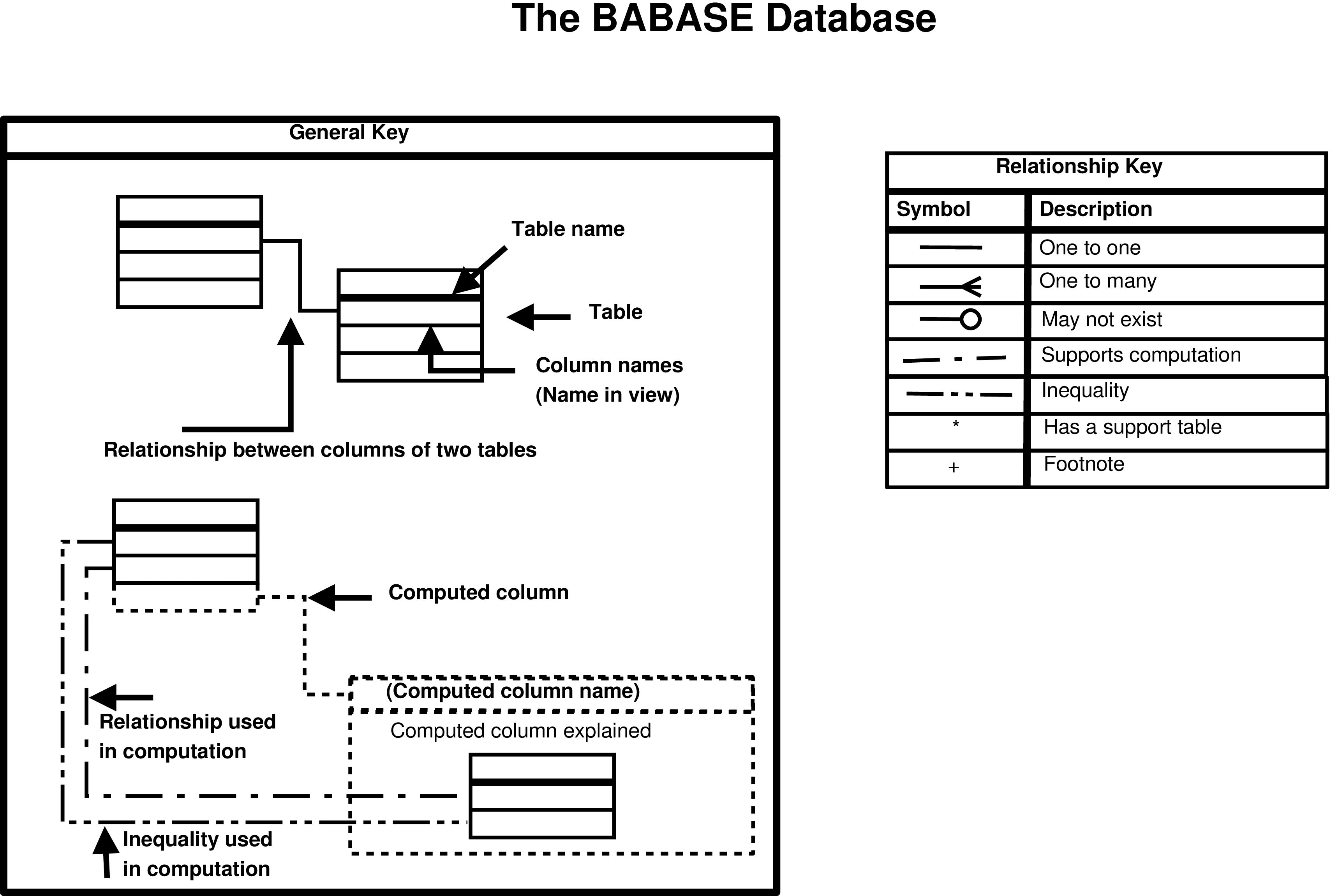
- 2.1. Key to the Babase Entity Relationship Diagrams
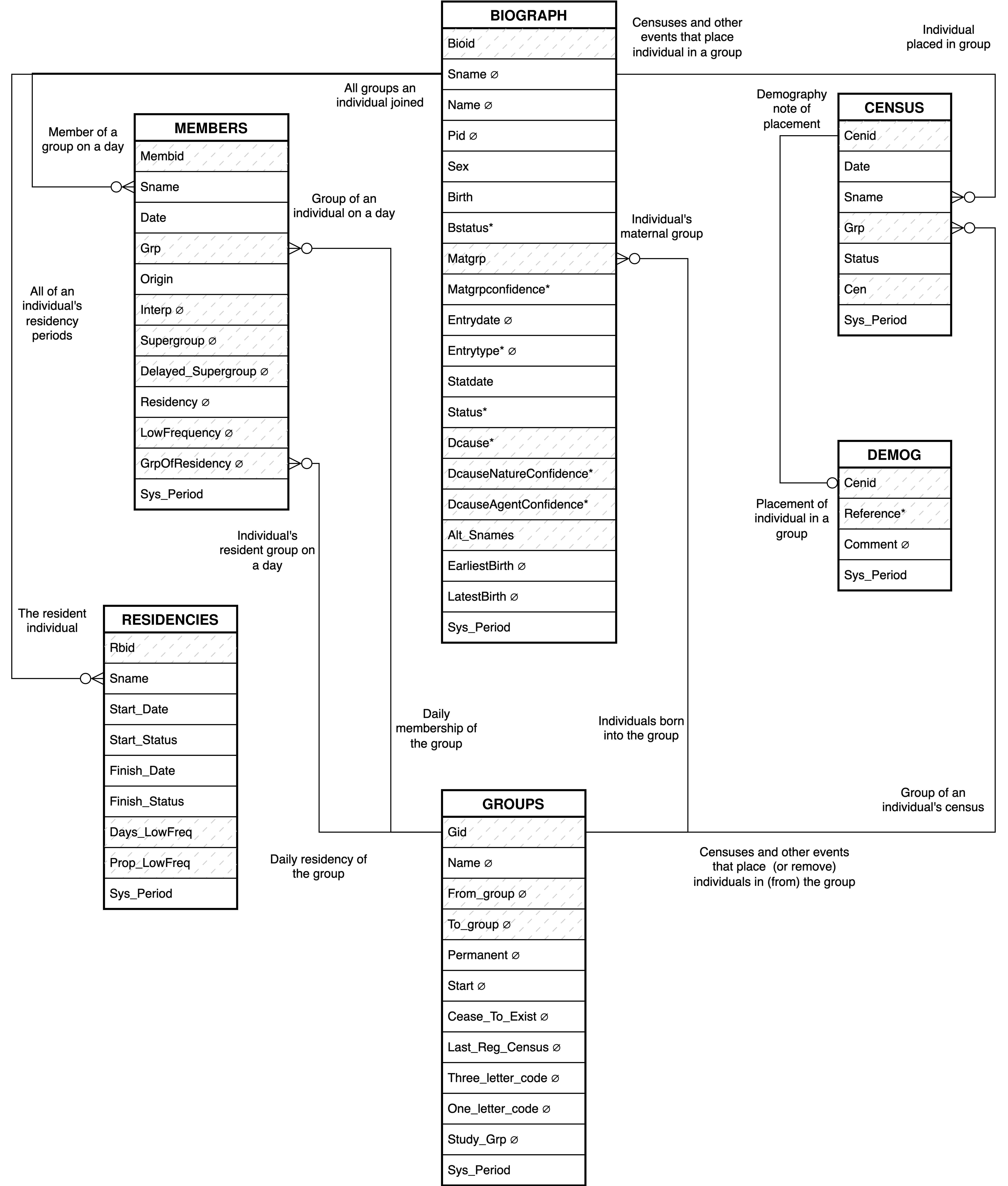
- 2.2. Babase Group Membership Entity Relationship Diagram
- 2.3. Babase Life Events Entity Relationship Diagram
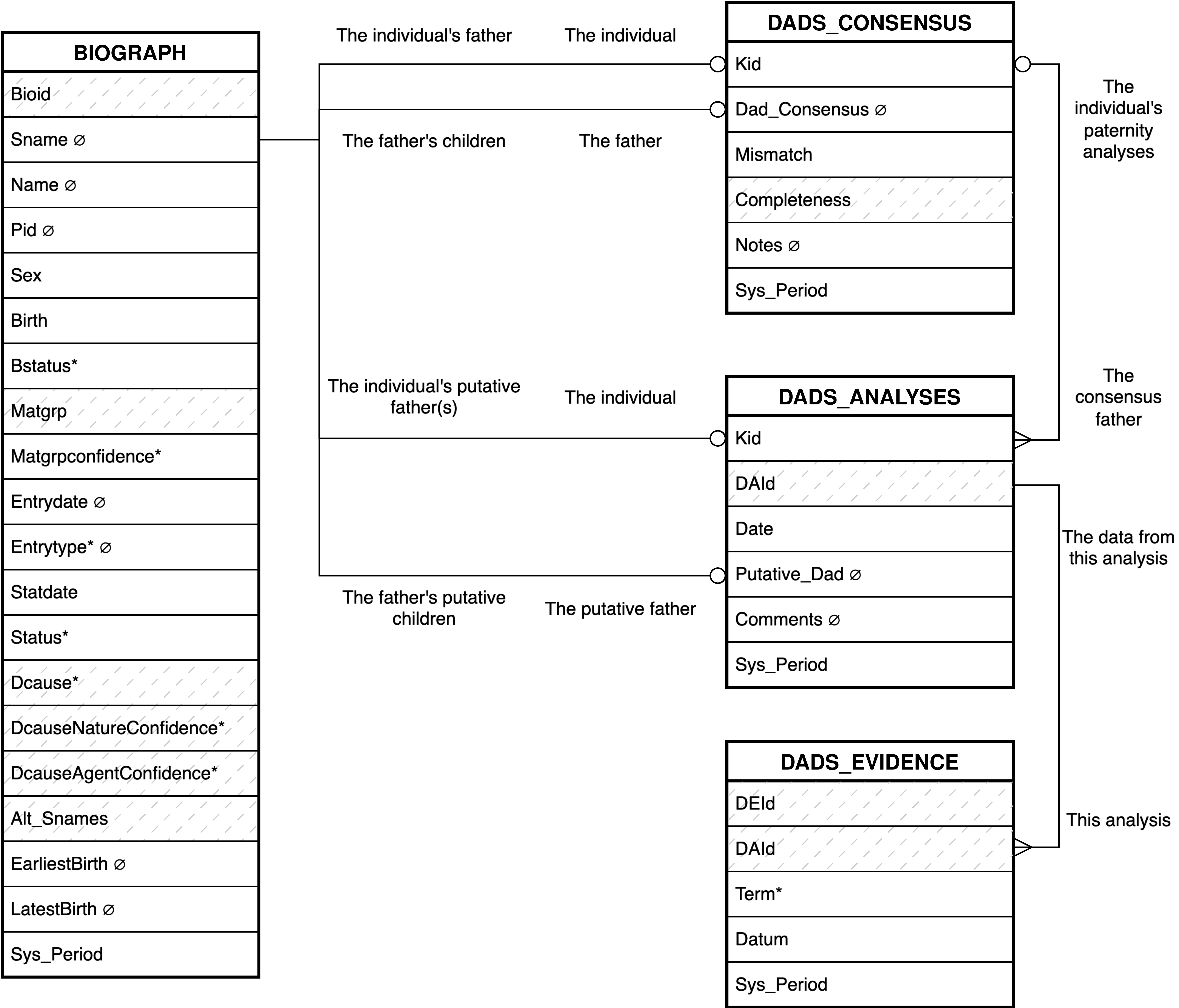
- 2.4. Babase Paternity Entity Relationship Diagram
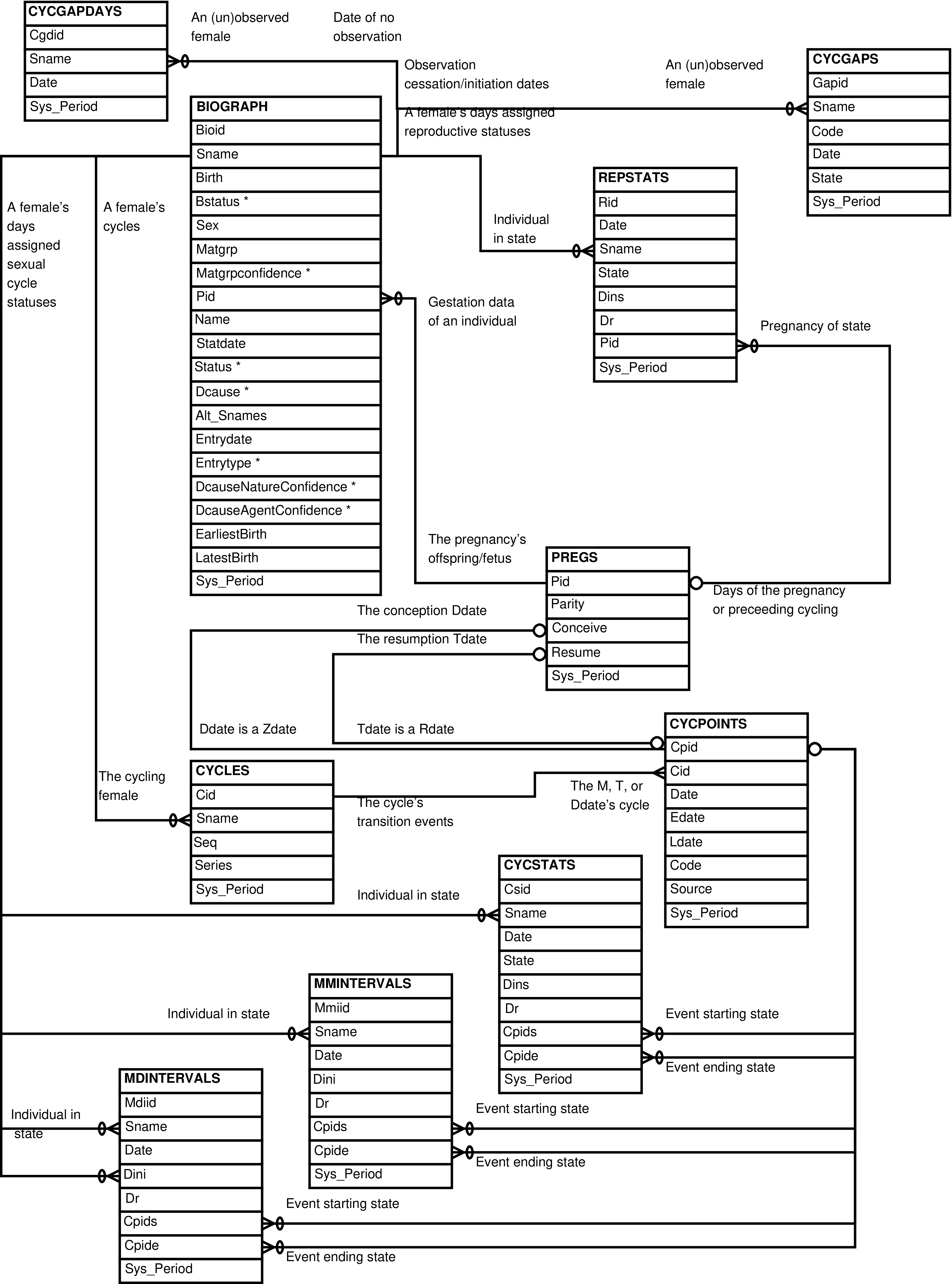
- 2.5. Babase Sexual Cycle Entity Relationship Diagram
- 2.6. Babase Sexual Cycle Day-To-Day Tables Entity Relationship Diagram
- 2.7. Babase Social Interactions Entity Relationship Diagram
- 2.8. Babase Multiparty Interactions Entity Relationship Diagram
- 2.9. Babase Darting Logistics and Morphology Entity and Relationship Diagram
- 2.10. Babase Darting Physiology Entity and Relationship Diagram
- 2.11. Babase Darting Samples Entity and Relationship Diagram
- 2.12. Babase Darting Teeth and Ticks Entity and Relationship Diagram
- 2.13. Babase Inventory Entity Relationship Diagram
- 2.14. Babase Physical Traits Hormone Data Entity Relationship Diagram
- 2.15. Babase Physical Traits Hybrid Score Data Entity Relationship Diagram
- 2.16. Babase Physical Traits Wounds and Pathologies Data Entity Relationship Diagram
- 2.17. Babase SWERB Core Tables Entity Relationship Diagram
- 2.18. Babase SWERB Grove/Waterhole Location Tables Entity Relationship Diagram
- 2.19. Babase Manual Weather Data Entity Relationship Diagram
- 2.20. Babase Digital Weather Data Entity Relationship Diagram
- 4.1. An Individual is Censused Present and Absent
- 4.2. Interpolating From Presences and Absences
- 4.3. Interpolating Group Membership
- 4.4. Computing Interp Values
- 4.5. The 14 Day Interpolation Limit
- 4.6. Interpolation at Birth
- 4.7. Alive and Present When Last Censused
- 4.8. Alive and Absent in Last Census
- 4.9. Interpolation to Statdate When Dead
- 4.10. Midpoint Days
- 4.11. The Midpoint Rule Adjusts Intervals
- 4.12. An Individual is Censused in 2 Groups
- 4.13. Interpolating Each Group Separately
- 4.14. A Closer Look at Intervals
- 4.15. Presence and Absence Interpolated Separately
- 4.16. Combining Presence and Absence Intervals
- 4.17. Group Membership Given Multiple Groups
- 4.18. Pre-Analyzed Data Truncates Interpolation Intervals
- 4.19. Pre-Analyzed Data Interrupts Interpolation
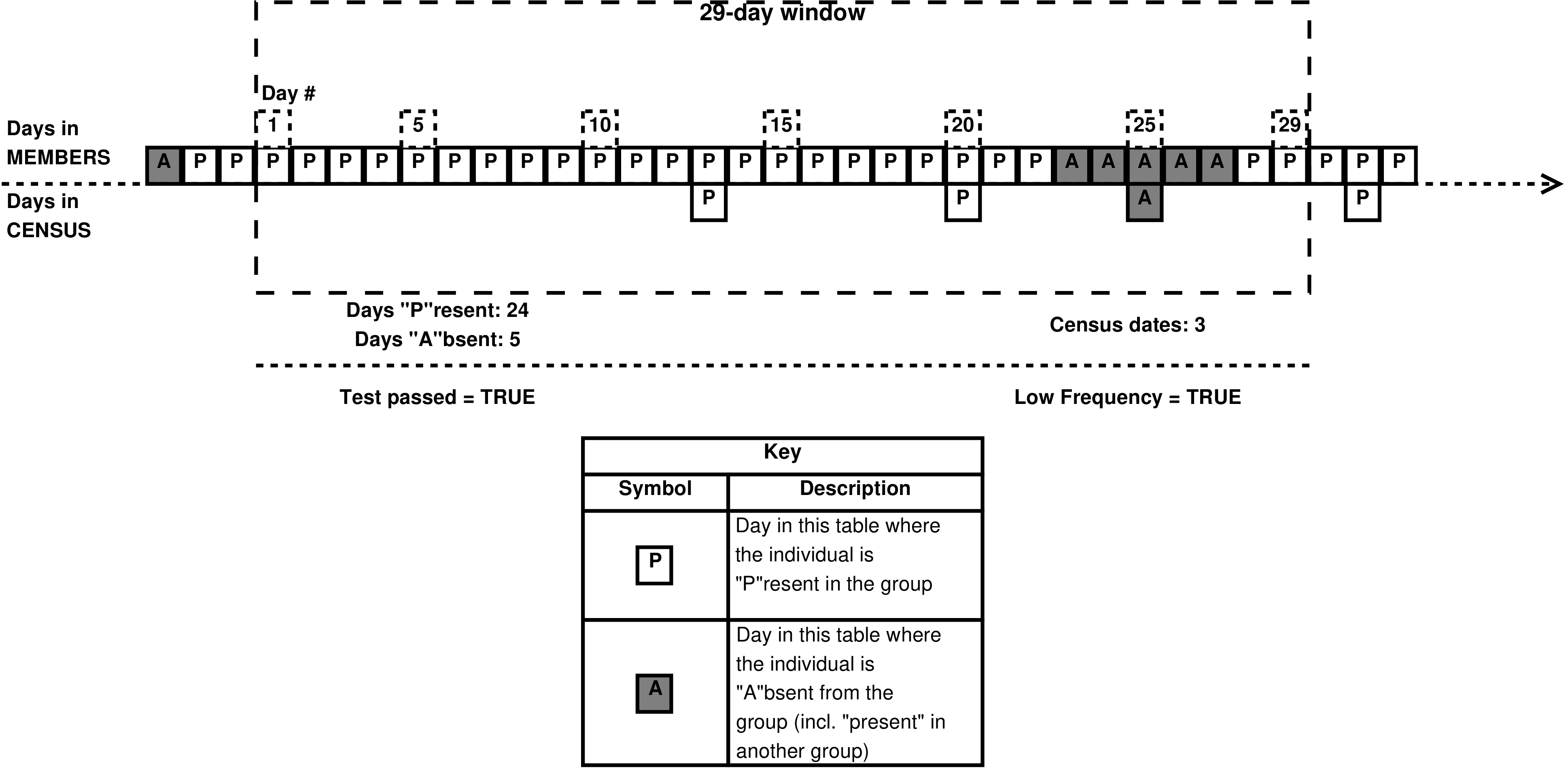
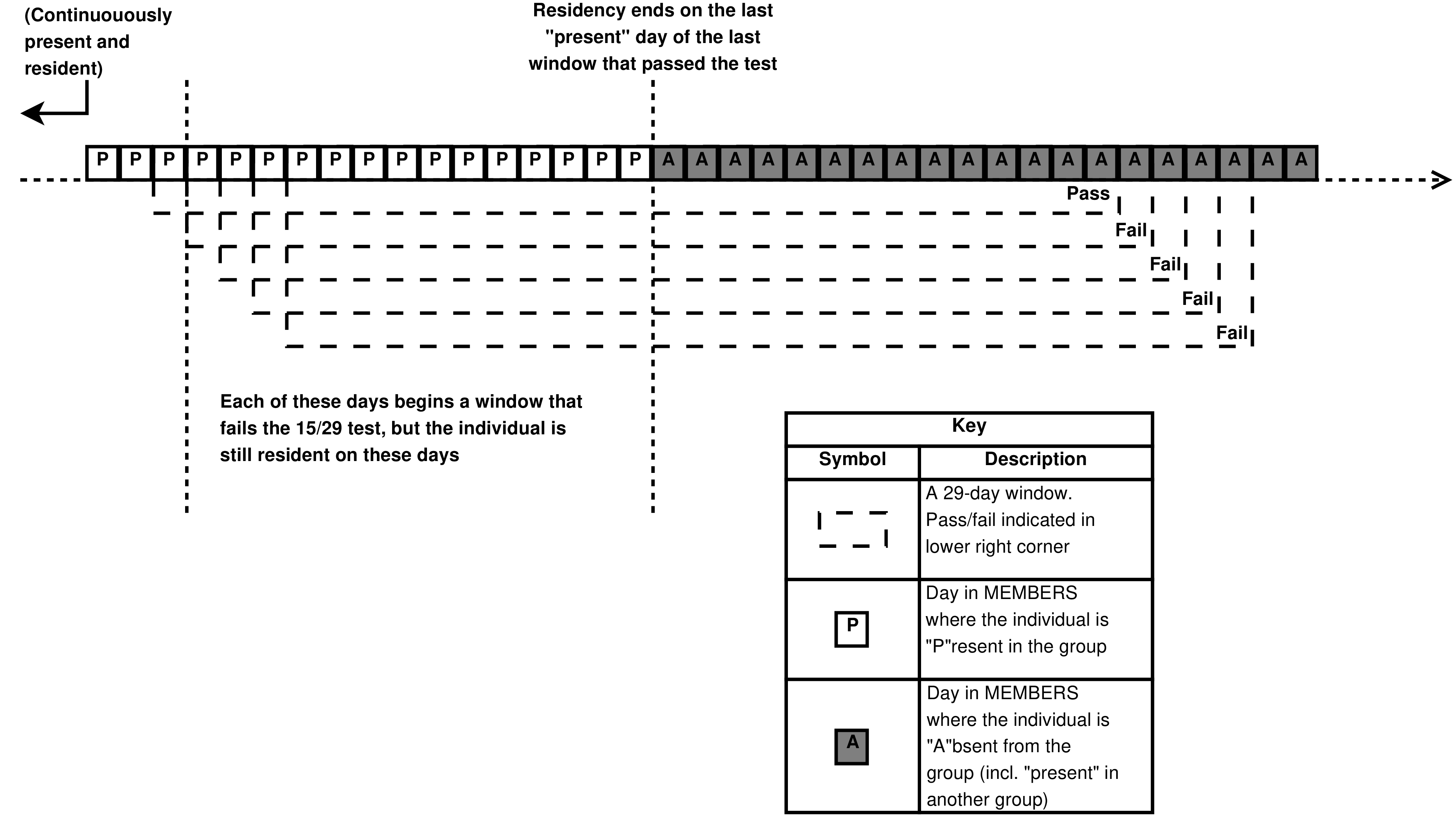
- 4.20. An example 15/29 test
- 4.21. 29-day windows at the end of a residency period
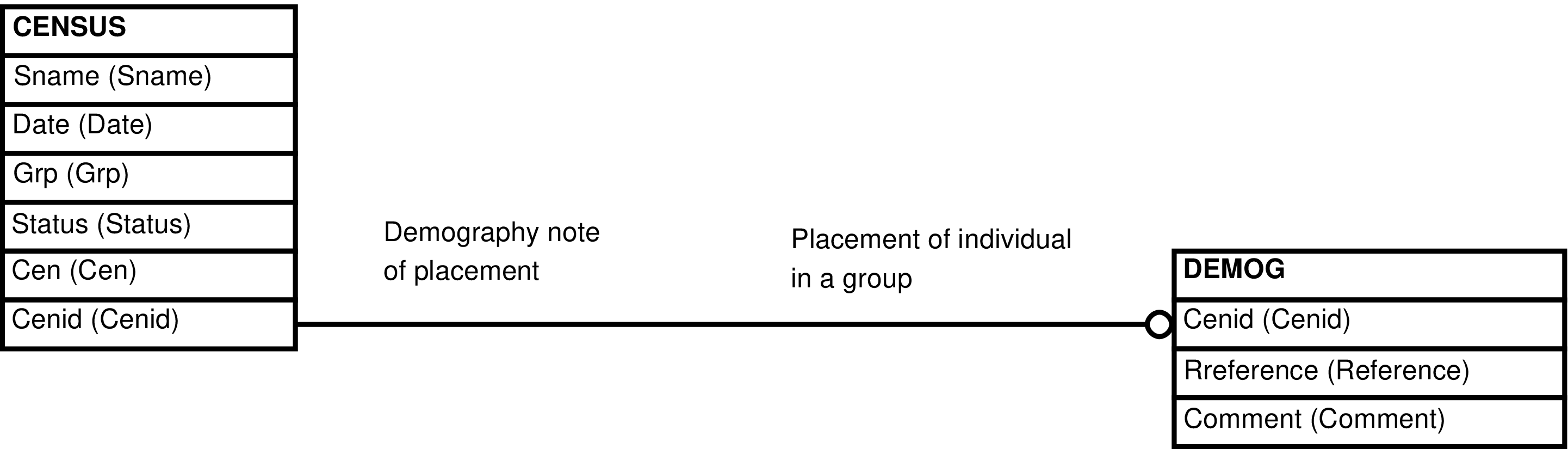
- 6.1. Query Defining the CENSUS_DEMOG View
- 6.2. Entity Relationship Diagram of the CENSUS_DEMOG View
- 6.3. Query Defining the CENSUS_DEMOG_SORTED View
- 6.4. Entity Relationship Diagram of the CENSUS_DEMOG_SORTED View
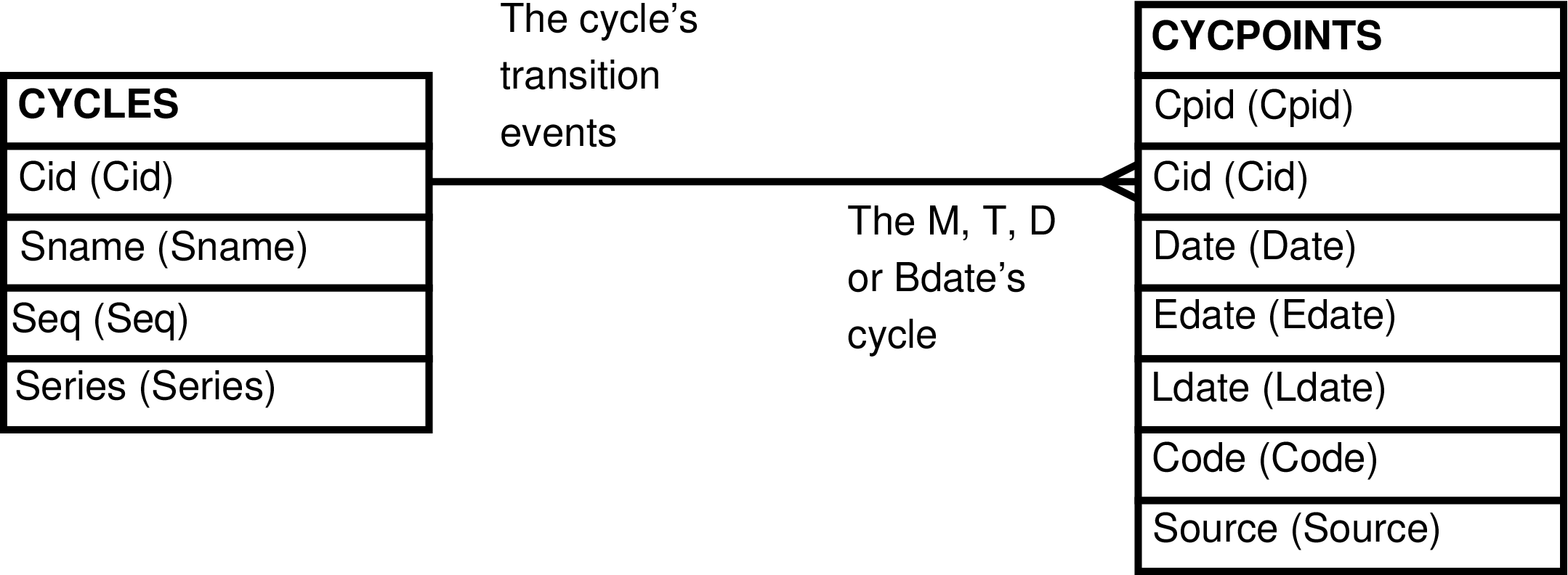
- 6.5. Query Defining the CYCPOINTS_CYCLES View
- 6.6. Entity Relationship Diagram of the CYCPOINTS_CYCLES View
- 6.7. Query Defining the CYCPOINTS_CYCLES_SORTED View
- 6.8. Entity Relationship Diagram of the CYCPOINTS_CYCLES_SORTED View
- 6.9. Query Defining the DEMOG_CENSUS View
- 6.10. Entity Relationship Diagram of the DEMOG_CENSUS View
- 6.11. Query Defining the DEMOG_CENSUS_SORTED View
- 6.12. Entity Relationship Diagram of the DEMOG_CENSUS_SORTED View
- 6.13. Query Defining the GROUPS_HISTORY View
- 6.14. Entity Relationship Diagram of the GROUPS_HISTORY View
- 6.15. Query Defining the PARENTS View
- 6.16. Entity Relationship Diagram of the PARENTS View
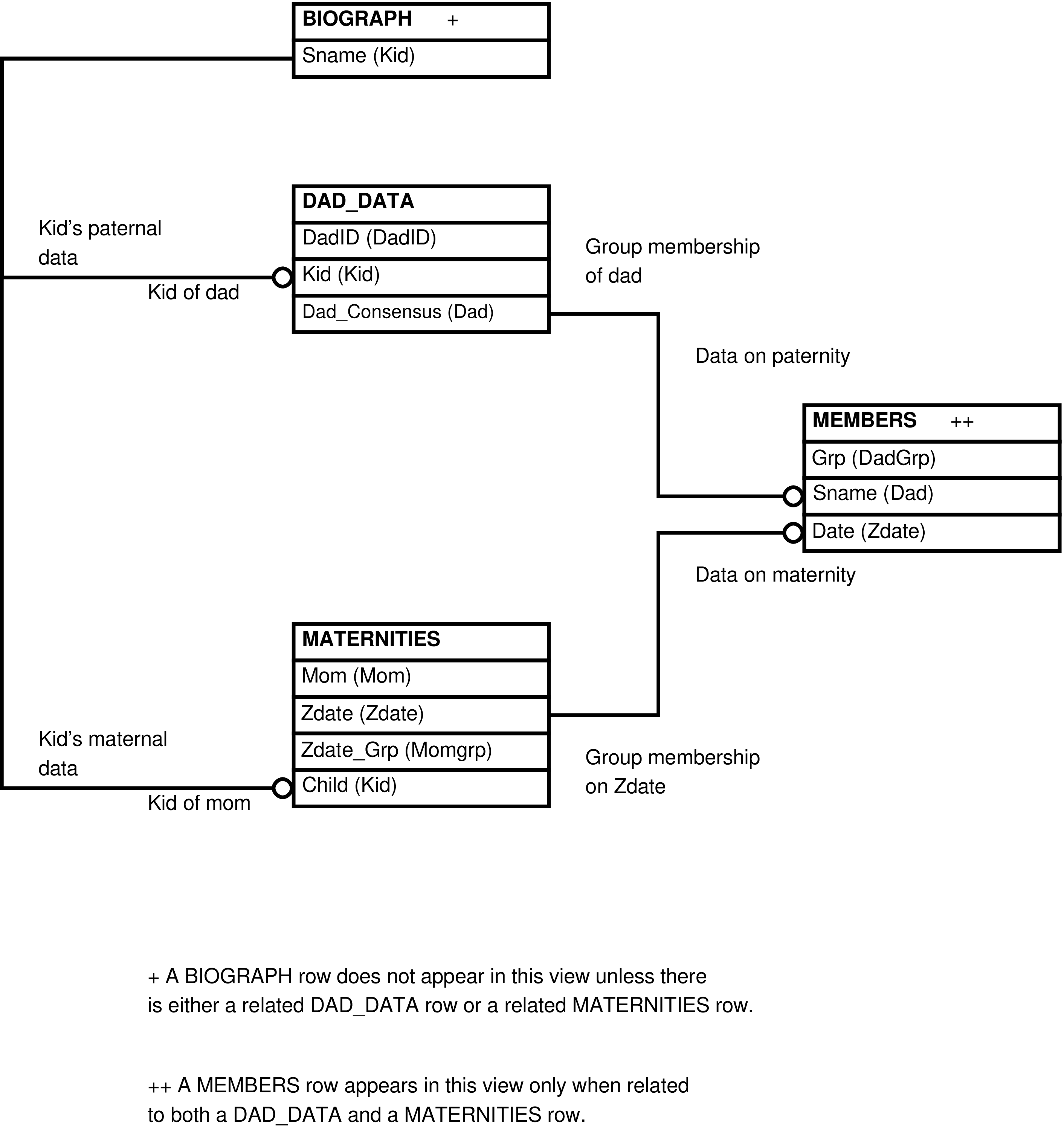
- 6.17. Query Defining the POTENTIAL_DADS View
- 6.18. Entity Relationship Diagram of the foundation of the POTENTIAL_DADS View
- 6.19. Entity Relationship Diagram of that portion of the POTENTIAL_DADS View which places the mother and potential father in the same group during the fertile period
- 6.20. Entity Relationship Diagram of that portion of the POTENTIAL_DADS View having easily computed columns
- 6.21. Entity Relationship Diagram of that portion of the POTENTIAL_DADS View involving social interactions
- 6.22. Query Defining the PROPORTIONAL_RANKS View
- 6.23. Entity Relationship Diagram of the PROPORTIONAL_RANKS View
- 6.24. Query Defining the ESTROGENS View
- 6.25. Entity Relationship Diagram of the ESTROGENS View
- 6.26. Query Defining the GLUCOCORTICOIDS View
- 6.27. Entity Relationship Diagram of the GLUCOCORTICOIDS View
- 6.28. Query Defining the HORMONE_PREPS View
- 6.29. Entity Relationship Diagram of the HORMONE_PREPS View
- 6.30. Query Defining the HORMONE_RESULTS View
- 6.31. Entity Relationship Diagram of the HORMONE_RESULTS View
- 6.32. Query Defining the HORMONE_SAMPLES View
- 6.33. Entity Relationship Diagram of the HORMONE_SAMPLES View
- 6.34. Query Defining the HYBRIDMORPH_SCORES View
- 6.35. Entity Relationship Diagram of the HYBRIDMORPH_SCORES View
- 6.36. Query Defining the PROGESTERONES View
- 6.37. Entity Relationship Diagram of the PROGESTERONES View
- 6.38. Query Defining the TESTOSTERONES View
- 6.39. Entity Relationship Diagram of the TESTOSTERONES View
- 6.40. Query Defining the THYROID_HORMONES View
- 6.41. Entity Relationship Diagram of the THYROID_HORMONES View
- 6.42. Query Defining the WOUNDSPATHOLOGIES View
- 6.43. Entity Relationship Diagram of the WOUNDSPATHOLOGIES View
- 6.44. Query Defining the WP_DETAILS_AFFECTEDPARTS View
- 6.45. Entity Relationship Diagram of the WP_DETAILS_AFFECTEDPARTS View
- 6.46. Query Defining the WP_HEALS View
- 6.47. Entity Relationship Diagram of the WP_HEALS View, Overall
- 6.48. Entity Relationship Diagram of the WP_HEALS View for rows with an update to a wound/pathology report
- 6.49. Entity Relationship Diagram of the WP_HEALS View for rows with an update to a wound/pathology cluster
- 6.50. Entity Relationship Diagram of the WP_HEALS View for rows with an update to an affected body part
- 6.51. Query Defining the WP_REPORTS_OBSERVERS View
- 6.52. Entity Relationship Diagram of the WP_REPORTS_OBSERVERS View
- 6.53. Query Defining the CYCLES_SEXSKINS View
- 6.54. Entity Relationship Diagram of the CYCLES_SEXSKINS View
- 6.55. Query Defining the CYCLES_SEXSKINS_SORTED View
- 6.56. Entity Relationship Diagram of the CYCLES_SEXSKINS_SORTED View
- 6.57. Query Defining the MATERNITIES View
- 6.58. Entity Relationship Diagram of the MATERNITIES View
- 6.59. Query Defining the MTD_CYCLES View
- 6.60. Entity Relationship Diagram of the MTD_CYCLES View
- 6.61. Query Defining the SEXSKINS_CYCLES View
- 6.62. Entity Relationship Diagram of the SEXSKINS_CYCLES View
- 6.63. Query Defining the SEXSKINS_CYCLES_SORTED View
- 6.64. Entity Relationship Diagram of the SEXSKINS_CYCLES_SORTED View
- 6.65. Query Defining the SEXSKINS_REPRO_NOTES View
- 6.66. Entity Relationship Diagram of the SEXSKINS_REPRO_NOTES View
- 6.67. Query Defining the ACTOR_ACTEES View
- 6.68. Entity Relationship Diagram of the ACTOR_ACTEES View
- 6.69. Query Defining the INTERACT View
- 6.70. Entity Relationship Diagram of the INTERACT View
- 6.71. Query Defining the INTERACT_SORTED View
- 6.72. Entity Relationship Diagram of the INTERACT_SORTED View
- 6.73. Query Defining the MPI_EVENTS View
- 6.74. Entity Relationship Diagram of the MPI_EVENTS View
- 6.75. Query Defining the MPI_UPLOAD View
- 6.76. Entity Relationship Diagram of the MPI_UPLOAD View
- 6.77. Query Defining the POINTS View
- 6.78. Entity Relationship Diagram of the POINTS View
- 6.79. Query Defining the POINTS_SORTED View
- 6.80. Entity Relationship Diagram of the POINTS_SORTED View
- 6.81. Query Defining the SAMPLES_GOFF View
- 6.82. Entity Relationship Diagram of the SAMPLES_GOFF View
- 6.83. Query Defining the ANESTH_STATS View
- 6.84. Entity Relationship Diagram of the ANESTH_STATS View
- 6.85. Query Defining the BODYTEMP_STATS View
- 6.86. Entity Relationship Diagram of the BODYTEMP_STATS View
- 6.87. Query Defining the CHEST_STATS View
- 6.88. Entity Relationship Diagram of the CHEST_STATS View
- 6.89. Query Defining the CROWNRUMP_STATS View
- 6.90. Entity Relationship Diagram of the CROWNRUMP_STATS View
- 6.91. Query Defining the DART_FLOW_CYTOMETRY_UPLOAD View
- 6.92. Entity Relationship Diagram of the DART_FLOW_CYTOMETRY_UPLOAD View
- 6.93. Query Defining the DART_LOGISTICS_UPLOAD View
- 6.94. Entity Relationship Diagram of the DART_LOGISTICS_UPLOAD View
- 6.95. Query Defining the DART_MORPHOLOGY_UPLOAD View
- 6.96. Entity Relationship Diagram of the DART_MORPHOLOGY_UPLOAD View
- 6.97. Query Defining the DART_PHYSIOLOGY_UPLOAD View
- 6.98. Entity Relationship Diagram of the DART_PHYSIOLOGY_UPLOAD View
- 6.99. Query Defining the DART_SAMPLES_UPLOAD View
- 6.100. Entity Relationship Diagram of the DART_SAMPLES_UPLOAD View
- 6.101. Query Defining the DART_TEETH_UPLOAD View
- 6.102. Entity Relationship Diagram of the DART_TEETH_UPLOAD View
- 6.103. Query Defining the DART_TESTES_ARC_UPLOAD View
- 6.104. Entity Relationship Diagram of the DART_TESTES_ARC_UPLOAD View
- 6.105. Query Defining the DART_TESTES_DIAM_UPLOAD View
- 6.106. Entity Relationship Diagram of the DART_TESTES_DIAM_UPLOAD View
- 6.107. Query Defining the DART_TICKS_UPLOAD View
- 6.108. Entity Relationship Diagram of the DART_TICKS_UPLOAD View
- 6.109. Query Defining the DART_VAGINAL_PHS_UPLOAD View
- 6.110. Entity Relationship Diagram of the DART_VAGINAL_PHS_UPLOAD View
- 6.111. Query Defining the DART_WBC_COUNTS_UPLOAD View
- 6.112. Entity Relationship Diagram of the DART_WBC_COUNTS_UPLOAD View
- 6.113. Query Defining the DSAMPLES View
- 6.114. Query Defining the DENT_CODES View
- 6.115. Entity Relationship Diagram of the DENT_CODES View
- 6.116. Query Defining the DENT_SITES View
- 6.117. Entity Relationship Diagram of the DENT_SITES View
- 6.118. Query Defining the HUMERUS_STATS View
- 6.119. Entity Relationship Diagram of the HUMERUS_STATS View
- 6.120. Query Defining the PCV_STATS View
- 6.121. Entity Relationship Diagram of the PCV_STATS View
- 6.122. Query Defining the TESTES_ARC_STATS View
- 6.123. Entity Relationship Diagram of the TESTES_ARC_STATS View
- 6.124. Query Defining the TESTES_DIAM_STATS View
- 6.125. Entity Relationship Diagram of the TESTES_DIAM_STATS View
- 6.126. Query Defining the ULNA_STATS View
- 6.127. Entity Relationship Diagram of the ULNA_STATS View
- 6.128. Query Defining the VAGINAL_PH_STATS View
- 6.129. Entity Relationship Diagram of the VAGINAL_PH_STATS View
- 6.130. Query Defining the LOCATIONS_FREE View
- 6.131. Entity Relationship Diagram of the LOCATIONS_FREE View
- 6.132. Query Defining the NUCACID_CONCS View
- 6.133. Entity Relationship Diagram of the NUCACID_CONCS View
- 6.134. Query Defining the NUCACID_SOURCES_EXT View
- 6.135. Entity Relationship Diagram of the NUCACID_SOURCES_EXT View
- 6.136. Query Defining the NUCACIDS View
- 6.137. Entity Relationship Diagram of the NUCACIDS View
- 6.138. Query Defining the NUCACIDS_W_CONC View
- 6.139. Entity Relationship Diagram of the NUCACIDS_W_CONC View
- 6.140. Query Defining the TISSUE_SOURCES_EXT View
- 6.141. Entity Relationship Diagram of the TISSUE_SOURCES_EXT View
- 6.142. Query Defining the TISSUES View
- 6.143. Entity Relationship Diagram of the TISSUES View
- 6.144. Query Defining the TISSUES_HORMONES View
- 6.145. Entity Relationship Diagram of the TISSUES_HORMONES View
- 6.146. Query Defining the QUADS View
- 6.147. Entity Relationship Diagram of the QUADS View
- 6.148. Query Defining the SWERB View
- 6.149. Entity Relationship Diagram of the SWERB View
- 6.150. Query Defining the SWERB_DATA_XY View
- 6.151. Entity Relationship Diagram of the SWERB_DATA_XY View
- 6.152. Query Defining the SWERB_DEPARTS View
- 6.153. Entity Relationship Diagram of the SWERB_DEPARTS View
- 6.154. Query Defining the SWERB_GW_LOCS View
- 6.155. Entity Relationship Diagram of the SWERB_GW_LOCS View
- 6.156. Query Defining the SWERB_GW_LOC_DATA_XY View
- 6.157. Entity Relationship Diagram of the SWERB_GW_LOC_DATA_XY View
- 6.158. Query Defining the SWERB_LOC_GPS_XY View
- 6.159. Entity Relationship Diagram of the SWERB_LOC_GPS_XY View
- 6.160. Query Defining the SWERB_LOCS View
- 6.161. Entity Relationship Diagram of the SWERB_LOCS View
- 6.162. Query Defining the SWERB_UPLOAD View
- 6.163. Entity Relationship Diagram of the SWERB_UPLOAD View
- 6.164. Query Defining the CURATED_TEMPS View
- 6.165. Entity Relationship Diagram of the portion of the CURATED_TEMPS View involving electronically-collected weather data
- 6.166. Entity Relationship Diagram of the portion of the CURATED_TEMPS View involving manually-collected weather data
- 6.167. Query Defining the MIN_MAXS View
- 6.168. Entity Relationship Diagram of the MIN_MAXS View
- 6.169. Query Defining the MIN_MAXS_SORTED View
- 6.170. Entity Relationship Diagram of the MIN_MAXS_SORTED View
- 6.171. Query Defining the BIRTH_GRP View
- 6.172. Entity Relationship Diagram of the BIRTH_GRP View
- 6.173. Query Defining the ENTRYDATE_GRP View
- 6.174. Entity Relationship Diagram of the ENTRYDATE_GRP View
- 6.175. Query Defining the STATDATE_GRP View
- 6.176. Entity Relationship Diagram of the STATDATE_GRP View
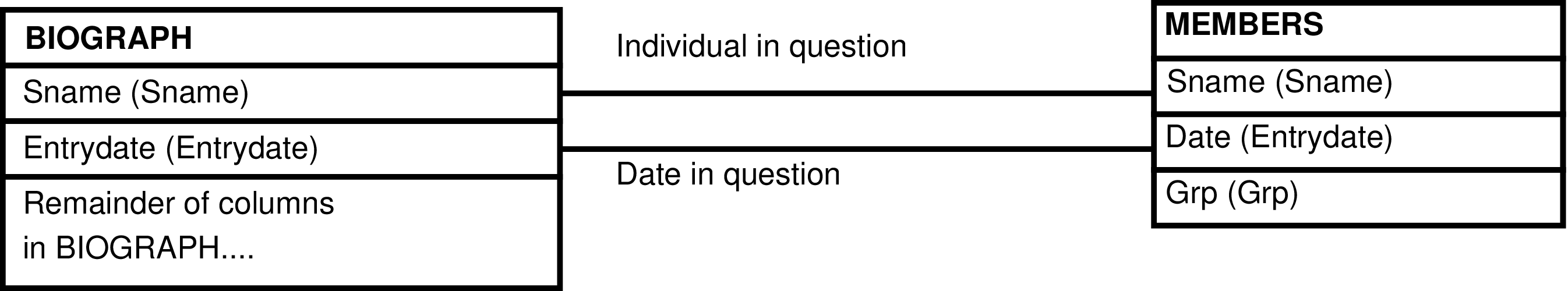
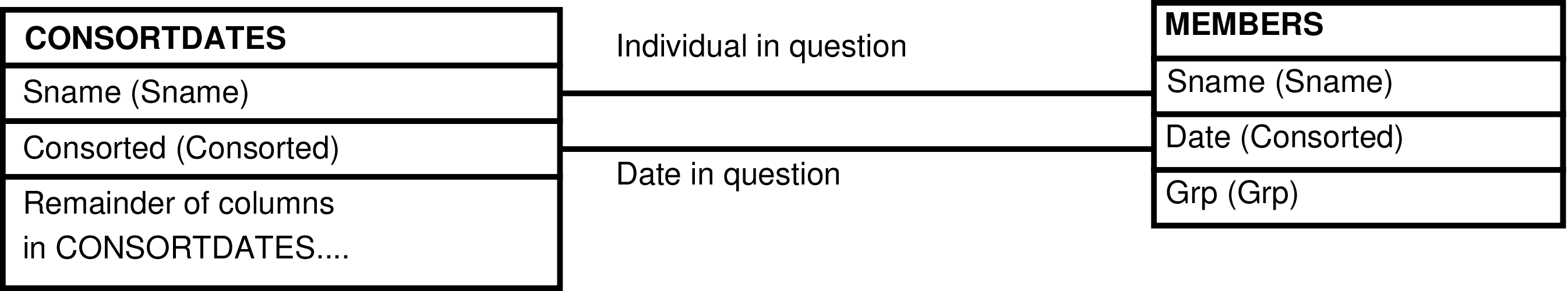
- 6.177. Query Defining the CONSORTDATES_GRP View
- 6.178. Entity Relationship Diagram of the CONSORTDATES_GRP View
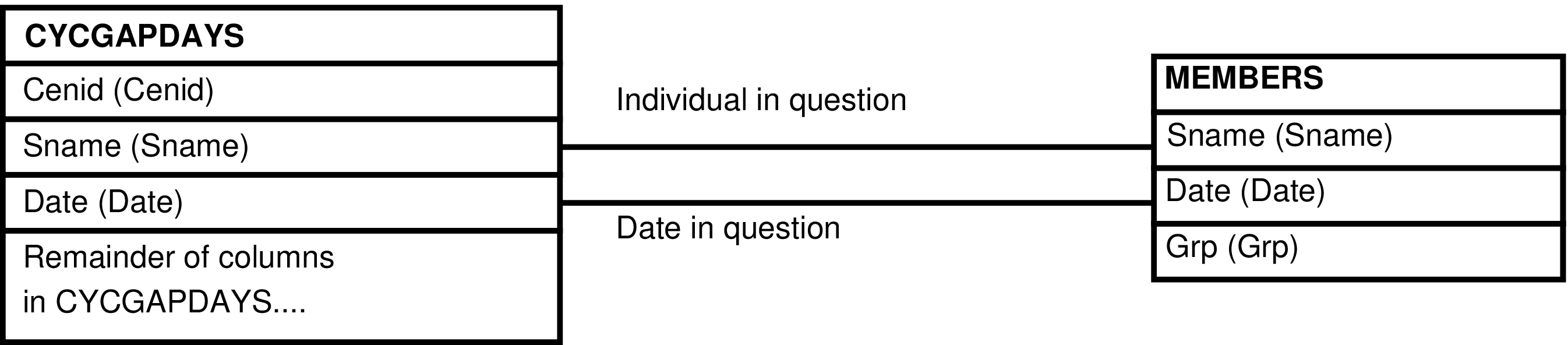
- 6.179. Query Defining the CYCGAPDAYS_GRP View
- 6.180. Entity Relationship Diagram of the CYCGAPDAYS_GRP View
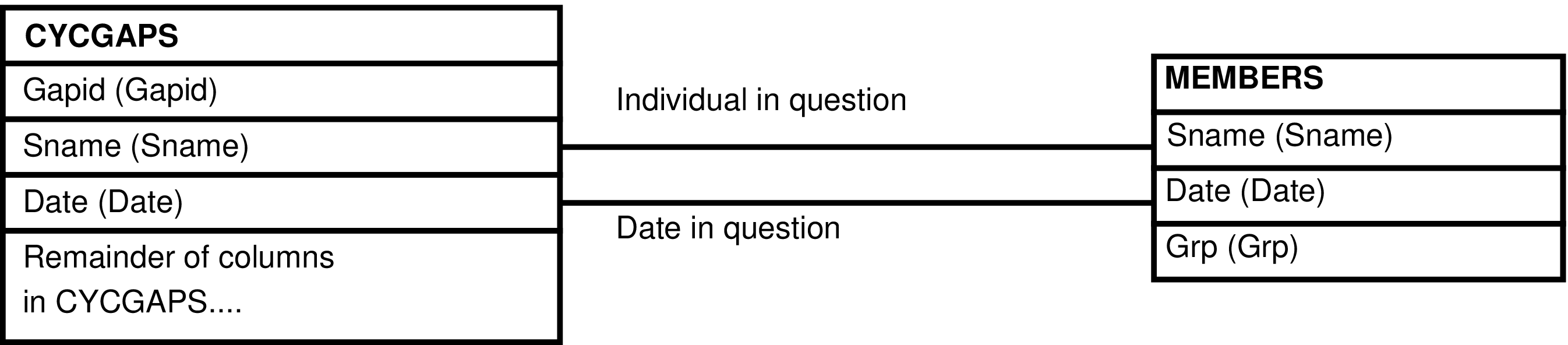
- 6.181. Query Defining the CYCGAPS_GRP View
- 6.182. Entity Relationship Diagram of the CYCGAPS_GRP View
- 6.183. Query Defining the CYCSTATS_GRP View
- 6.184. Entity Relationship Diagram of the CYCSTATS_GRP View
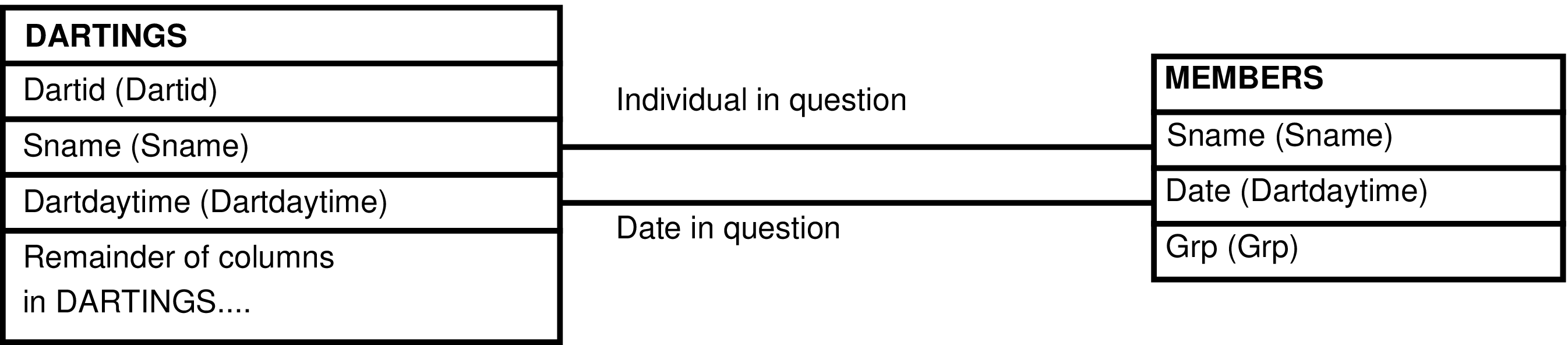
- 6.185. Query Defining the DARTINGS_GRP View
- 6.186. Entity Relationship Diagram of the DARTINGS_GRP View
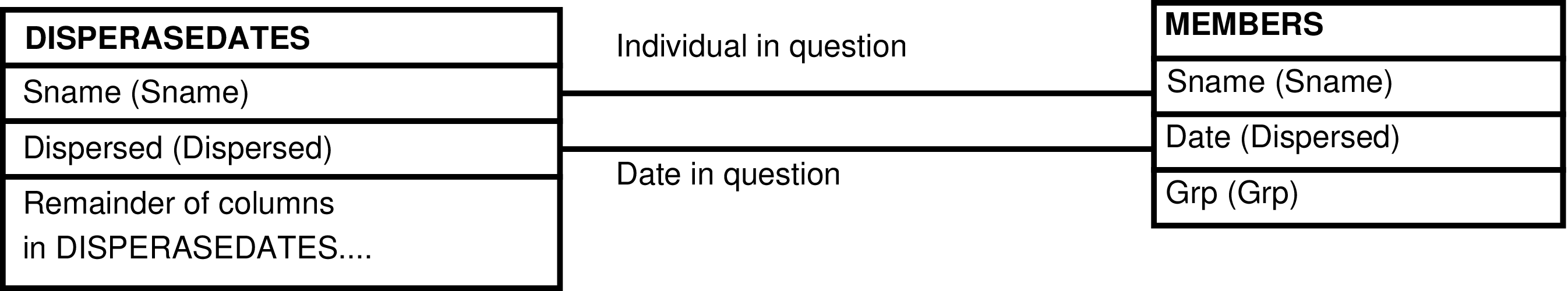
- 6.187. Query Defining the DISPERSEDATES_GRP View
- 6.188. Entity Relationship Diagram of the DISPERSEDATES_GRP View
- 6.189. Query Defining the MATUREDATES_GRP View
- 6.190. Entity Relationship Diagram of the MATUREDATES_GRP View
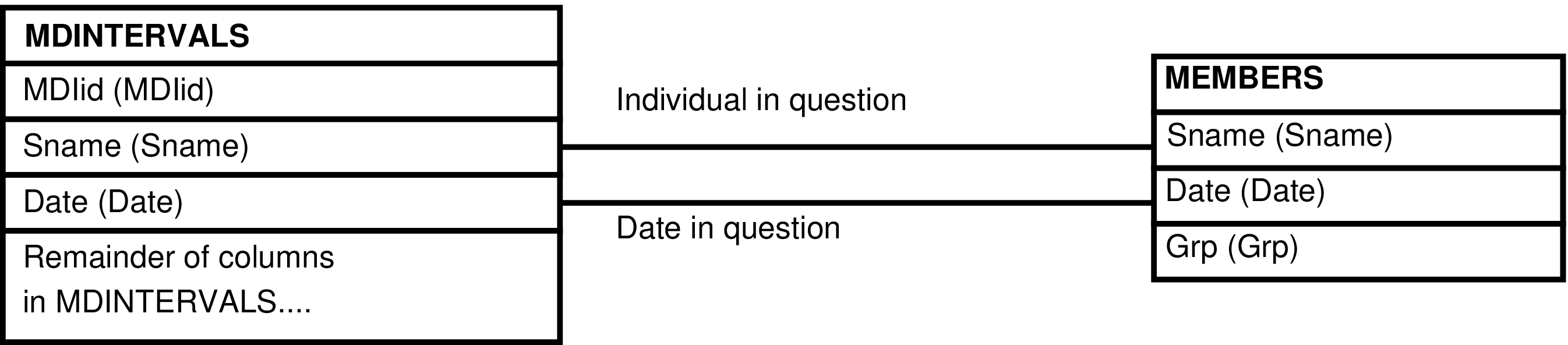
- 6.191. Query Defining the MDINTERVALS_GRP View
- 6.192. Entity Relationship Diagram of the MDINTERVALS_GRP View
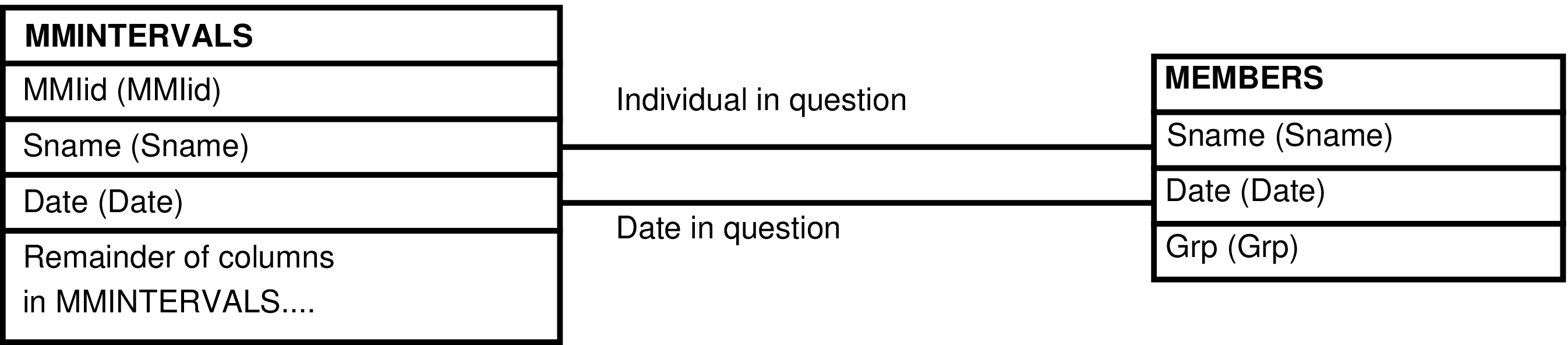
- 6.193. Query Defining the MMINTERVALS_GRP View
- 6.194. Entity Relationship Diagram of the MMINTERVALS_GRP View
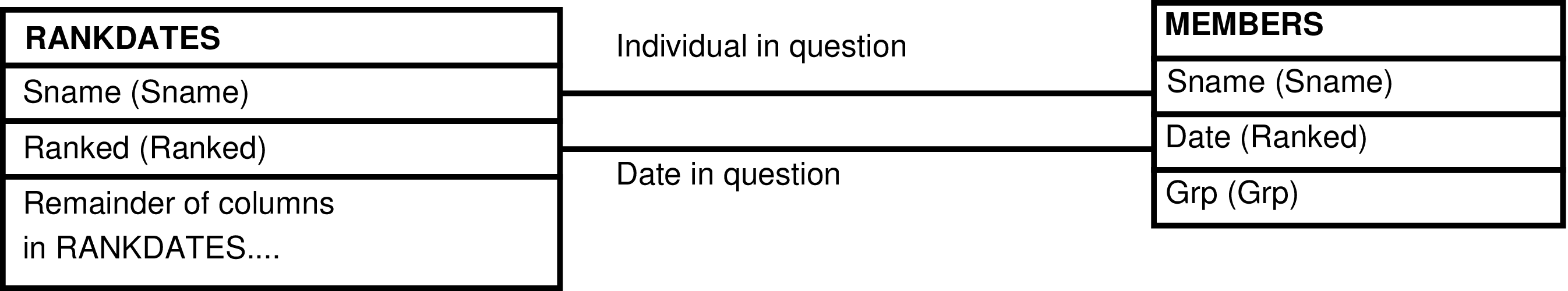
- 6.195. Query Defining the RANKDATES_GRP View
- 6.196. Entity Relationship Diagram of the RANKDATES_GRP View
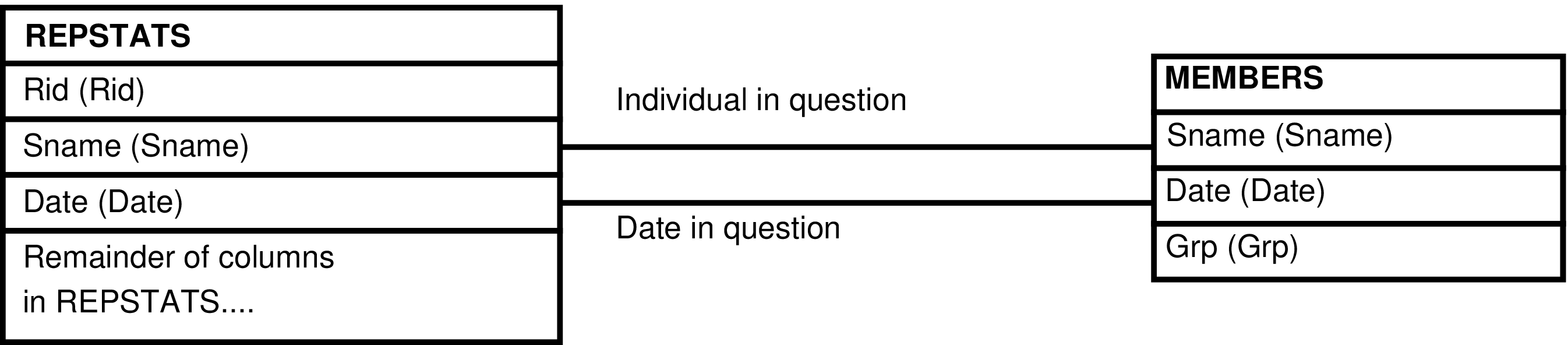
- 6.197. Query Defining the REPSTATS_GRP View
- 6.198. Entity Relationship Diagram of the REPSTATS_GRP View
List of Tables
- 2.2. The Main Babase Tables
- 2.3. The Babase Support Tables
- 2.4. The Babase Views
- 2.5. The table_GRP Views
- 6.1. Columns in the CENSUS_DEMOG View
- 6.2. Columns in the CENSUS_DEMOG_SORTED View
- 6.3. Columns in the CYCPOINTS_CYCLES View
- 6.4. Columns in the CYCPOINTS_CYCLES_SORTED View
- 6.5. Columns in the DEMOG_CENSUS View
- 6.6. Columns in the DEMOG_CENSUS_SORTED View
- 6.7. Columns in the GROUPS_HISTORY View
- 6.8. Columns in the PARENTS View
- 6.9. Columns in the POTENTIAL_DADS View
- 6.10. Columns in the PROPORTIONAL_RANKS View
- 6.11. Columns in the ESTROGENS View
- 6.12. Columns in the GLUCOCORTICOIDS View
- 6.13. Columns in the HORMONE_PREPS View
- 6.14. Columns in the HORMONE_RESULTS View
- 6.15. Columns in the HORMONE_SAMPLES View
- 6.16. Columns in the HYBRIDMORPH_SCORES View
- 6.17. Columns in the PROGESTERONES View
- 6.18. Columns in the TESTOSTERONES View
- 6.19. Columns in the THYROID_HORMONES View
- 6.20. Columns in the WOUNDSPATHOLOGIES View
- 6.21. Columns in the WP_DETAILS_AFFECTEDPARTS View
- 6.22. Columns in the WP_HEALS View
- 6.23. Columns in the WP_REPORTS_OBSERVERS View
- 6.24. Columns in the CYCLES_SEXSKINS View
- 6.25. Columns in the CYCLES_SEXSKINS_SORTED View
- 6.26. Columns in the MATERNITIES View
- 6.27. Columns in the MTD_CYCLES View
- 6.28. Columns in the SEXSKINS_CYCLES View
- 6.29. Columns in the SEXSKINS_CYCLES_SORTED View
- 6.30. Columns in the SEXSKINS_REPRO_NOTES View
- 6.31. Columns in the ACTOR_ACTEES View
- 6.32. Columns in the INTERACT View
- 6.33. Columns in the INTERACT_SORTED View
- 6.34. Columns in the MPI_EVENTS View
- 6.35. Columns in the POINTS View
- 6.36. Columns in the POINTS_SORTED View
- 6.37. Columns in the SAMPLES_GOFF View
- 6.38. Columns in the ANESTH_STATS View
- 6.39. Columns in the BODYTEMP_STATS View
- 6.40. Columns in the CHEST_STATS View
- 6.41. Columns in the CROWNRUMP_STATS View
- 6.42. Columns in the DART_FLOW_CYTOMETRY_UPLOAD View
- 6.43. Columns in the DART_LOGISTICS_UPLOAD View
- 6.44. Columns in the DART_MORPHOLOGY_UPLOAD View
- 6.45. Columns in the DART_PHYSIOLOGY_UPLOAD View
- 6.46. Columns in the DART_SAMPLES_UPLOAD View
- 6.47. Columns in the DART_TEETH_UPLOAD View
- 6.48. Columns in the DART_TESTES_ARC_UPLOAD View
- 6.49. Columns in the DART_TESTES_DIAM_UPLOAD View
- 6.50. Columns in the DART_TICKS_UPLOAD View
- 6.51. Columns in the DART_VAGINAL_PHS_UPLOAD View
- 6.52. Columns in the DART_WBC_COUNTS_UPLOAD View
- 6.53. Columns in the DSAMPLES View
- 6.54. Columns in the DENT_CODES View
- 6.55. Columns in the DENT_SITES View
- 6.56. Columns in the HUMERUS_STATS View
- 6.57. Columns in the PCV_STATS View
- 6.58. Columns in the TESTES_ARC_STATS View
- 6.59. Columns in the TESTES_DIAM_STATS View
- 6.60. Columns in the ULNA_STATS View
- 6.61. Columns in the VAGINAL_PH_STATS View
- 6.62. Columns in the LOCATIONS_FREE View
- 6.63. Columns in the NUCACID_CONCS View
- 6.64. Columns in the NUCACID_SOURCES_EXT View
- 6.65. Columns in the NUCACIDS View
- 6.66. Columns in the NUCACIDS_W_CONC View
- 6.67. Columns in the TISSUE_SOURCES_EXT View
- 6.68. Columns in the TISSUES View
- 6.69. Columns in the TISSUES_HORMONES View
- 6.70. Columns in the QUADS View
- 6.71. Columns in the SWERB View
- 6.72. Columns in the SWERB_DATA_XY View
- 6.73. Columns in the SWERB_DEPARTS View
- 6.74. Columns in the SWERB_GW_LOCS View
- 6.75. Columns in the SWERB_GW_LOC_DATA_XY View
- 6.76. Columns in the SWERB_LOC_GPS_XY View
- 6.77. Columns in the SWERB_LOCS View
- 6.78. Columns in the SWERB_UPLOAD View
- 6.79. Columns in the CURATED_TEMPS View
- 6.80. Columns in the MIN_MAXS View
- 6.81. Columns in the MIN_MAXS_SORTED View
- 8.1. The Babase SQL Functions
- 8.2. Data Analysis Procedures
List of Examples
- 1.1. A note
- 1.2. A caution
- 1.3. A warning
- 1.4. Text denoted important
- 1.5. A tip
- 2.1. Creating table
fooin the sandbox schema - 2.2. Granting permission to table
fooin the sandbox schema - 2.3. Creating table
fooin usermylogin's schema - 3.1. Crossovers during a fusion
- 4.1. An Agonism Matrix
- 4.2. Kits with no Correction, and NULL Correction
- 4.3. A familiar "series" of events
- 4.4. Determining the GrpOfResidency when absent
- 4.5. Resident in a nonexistent group
- A.1. Using the Postgresql
date_trunc()function to set seconds to zero - A.2. Using the Babase
date_mod()function to return the minutes and seconds. - A.3. Using the Postgresql
to_char()function to convert times to HH:MM text - B.1. Finding all the all-occurrences interactions
- C.1. “Splitting” a sexual cycle in two
- I.1. Querying "as of" a date
- I.2. Querying a table's history "as of" a date
Table of Contents
This document describes the Babase baboon data management system. This includes a description of the tables, the intended use of all related programs and directories, the design of the system, and procedures for maintaining the data management system itself. This document does not include the procedures actually used to enter data into the system, or the details of how to operate the systems programs. Nor does it include any instructions on the operation or administration of the computer itself. Further information on the topics not covered in this document can be found in the Protocol for Data Management: Amboseli Baboon Project document.
The Protocol for Data Management: Amboseli Baboon Project document is an important adjunct to the Babase system, but it is not considered part of the system itself because it describes the use of the system but not the capabilities of the system. It is important to maintain the distinction between use and capabilities so that when an enhancement is needed, it is clear whether the desired result can be obtained by altering the way the system is used, or whether the system itself needs to be modified. It is also important to provide different types of documentation to those who operate the system from those who manage and maintain the system because each of these two groups do not need to know all the details of the others' work.
Any deviation from the standards described in this document should be discussed with the project directors and may God have mercy on your souls.
This document follows a number of conventions, most of them typographic but some of them stylistic. Some output formats, particularly plain text, have limited typographic capabilities so the various forms of typographic markup are not always distinguishable, either from each other or from the surrounding text.
Each table in Babase is documented in a section of its own, beginning with a description of the table as a whole and continuing with sub-sections for each column in the table. Of particular importance is the sentence that describes what a row in the given table represents. These are summarized in the textual tables given in the Table Overview section.
Interrelationships between the columns of a table, or between tables, is documented at the beginning of the table's section, not in the sub-sections documenting the columns themselves. Although relationships between 2 tables concern both of the tables the description of each such relationship appears only once in this document, in the overall description of one of the of the two tables concerned. On occasion there may be be brief mention elsewhere.
All TABLE NAMES are written in UPPER CASE. Column Names are in lower case with Initial Capitals. SOMETABLE.Somecolumn is shorthand for “the Somecolumn column of the SOMETABLE table”. The use of a period to separate the table from the column name is the convention used by SQL to eliminate ambiguity regarding which table a column belongs to. When a column name includes an acronym the acronym is capitalized, as is the first letter of the next word when the acronym begins the column name. For example, PCSColor.
Actual database values are typographically distinguished
from the surrounding text, as in the following sentence:
“The Sname (short name) of the baboon Pebbles is
PEB.”
When this document defines a word, uses it for the first time, or otherwise wishes to refer to a word or phrase as a thing in and of itself, the word or phrase is typographically distinguished as follows: “The word census has several meanings within this document.”
Text that has special meaning to computer systems is
typographically distinguished as follows: “The SQL
SELECT statement is the standard method for
retrieving data from relational databases.”
Emphasized text is typographically distinguished as follows: “Always backup your data.”
When the words must or cannot or the phrases must not or may not are used, the system will not allow a contrary condition. For example: "Sname must be a unique data value" or "A user with read-only permissions may not change data values." Babase will immediately raise an error when a dis-allowed change is attempted and the change will not take effect.
When the words should or ought are used the system does not enforce the condition. It may or may not report a violation of the condition. An example: "The sexual cycle event referred to in the pregnancy table's Conceive column should date the conception that began the pregnancy." In this case the system has no way of knowing when the pregnancy began and so no way of validating the date.
When the phrase the system will report is used there is some mechanism for reporting a an unusual but not dis-allowed condition. Unlike prohibited conditions, unusual conditions are not generally reported at the time the condition is created.[1]
The documentation is written with a tendency to emphasize Special Values. So, for example, “not alive” is often written instead of “dead” because Babase has a special value that means alive but the system is not aware of a particular code that means dead. The result is an occasional double negative.
Significant but often slightly off-topic paragraphs are set off from the surrounding material as a note, shown in Example 1.1.
Example 1.1. A note
Note
Written material has no voice that can be raised, but attention can be drawn with typographical conventions.
When the reader should take care, particularly when the system might do something unexpected in a given circumstance, this is noted in a caution. Example 1.2 shows how a caution is set off from the surrounding text.
Example 1.2. A caution
Babase will reject your change if you try to do something that is not allowed, like giving a male an onset of turgesence date.
Caution
When the rejected change is one of a number of changes bundled into a transaction none of the changes will make it into the database.
When a mis-use of the system will lead to incorrect results, particularly when such results are not obvious, this document contains a warning. Example 1.3 show how warnings are set off from the surrounding text.
Example 1.3. A warning
Warning
Babase cannot detect when an Sname is mis-typed, so it is possible to inadvertently assign a female's sexual cycle to the wrong female.
To otherwise draw the readers attention to material some text is marked important. Example 1.4 show how important text is set off from the surrounding material.
Example 1.4. Text denoted important
Babase has a number of components, many of them, like the SQL web interface, are third party tools, not written by the Babase developers.
Important
When the third party tools are upgraded their “look” may change but the features they provide should remain. As Babase is composed of Free Software the Babase project always has the option of customizing any of its third party tools and can contribute its improvements back to the program's developers for inclusion into future releases.
Suggestions as to how to use Babase are noted in tips, as are remarks on how data are presently entered in Babase or recorded in the field. Example 1.5 show how a tip is set off from the regular document text.
Often, the tips are the result of best practice
developed from considered experience and so document how
Babase is used at the time of this writing. However, as best
practice continues to develop and field protocols change, the
Protocol for Data
Management: Amboseli Baboon Project and the Amboseli
Baboon Research Project Monitoring Guide should always be
consulted. Those documents have precedence over the tips
presented herein should there be conflicting advice.
Supplemental and cross-referential material is presented in footnotes.
Anyone who is changing or adding programs to the system should read this entire document. Chapter 3: “Baboon Data: Primary Source Material” is particularly important for all those using the system. Chapter 2: “Babase System Architecture” provides the introduction to Babase. It explains fundamental concepts without which Babase cannot be understood, although some portions can be skipped; the sections “The Babase Program Code” and “Indexes” are primarily of interest to programmers and the section “Special Values” is for the data maintainers. Everyone will want to pay special attention to the “Entity-Relationship Diagrams” section. These diagrams can also be found in PDF form in The Babase Pocket Reference, where they may be easier on the eye. The section “Data Maintenance Programs and Views” of chapter 8: “Babase Programs” is of little interest to those who only want to retrieve information from the system. Portions of the “Useful Programs and Functions” section of the same chapter is of interest to the more sophisticated user. Note that some functions may be hidden in “Next” links, depending on the format chosen when reading this document. Data maintainers should be sure to understand chapter 5: “Support Tables”. Those who are only retrieving data from Babase need not read chapter 7: “Data Entry”.
The Babase system is designed to facilitate the retrieval, storage, and maintenance of the Amboseli Baboon Project data. Data integrity is foremost. Analytical power, ease of use, and low cost are secondary goals. The system consists of tables to store and organize the data, software supporting data validation and derivative data generation, stand-alone programs used to facilitate the entry and maintenance of the data, a minimal tool set supporting the maintenance of the Babase system software itself, and documentation. data are retrieved from Babase using the SQL language, the standard[2] language used to query relational databases. SQL is declarational as opposed to procedural; from a single SQL query (a single statement) the database determines how to best retrieve the data requested, no matter the number of tables or criteria required. SQL provides a single, powerful, interface for ad-hoc data retrieval and manipulation. Generic software provides the bulk of the user interface[3], traditionally the most complex and costly software component.[4] Consequently there are few stand-alone programs written specifically for Babase. The overall philosophy of the systems implementation is to keep the software as easy to maintain as possible while assuring data integrity. To this end, the system is comprised of as many generic components as possible and the design requires custom programming for only the most crucial features.
Babase puts as much intelligence as possible into the database itself, including automatic data validation and complex automatic analysis and storage of the derived data.[5] Babase extends its sometimes complex and rather abstract database structures with alternative, more familiar and user-friendly, means of accessing the underlying data[6]. These constructs are, in so far as is possible, made indistinguishable from the underlying data when querying and updating the database. Babase often generates derivative data for more ready analysis. This is, for the most part, transparent to the user. The end-user is insulated from implementation details, the number of interfaces (primarily SQL) the user must learn is minimized, and the user is free to work with the data structures that embody the conceptual model best suited to the task at hand.[7]
Data input is an example of how Babase incorporates generic programs. The prototypical way to import data into Babase is in bulk, via a plain text file having columns delimited by the tab character. These are easily produced by almost any spreadsheet program; it is expected that most data imported into Babase will be typed into a spreadsheet and then exported to tab-delimited text for upload.[8] The use of generic interfaces reduces cost, and minimizing the number of novel interfaces frees the end-user to concentrate on the task at hand.
Babase is designed to be accessed over the Internet, primarily via the web. Although there are exceptions[9] the majority of Babase is accessed via a W3C compliant web browser. Individually assigned usernames and passwords are used, along with encryption, to secure the database content. The Babase Wiki provides content for an the structure of the project's web site. Another example of Babase leveraging a generic program, the wiki allows project members collaborate, share information, and build the project's web site without programmer intervention.
Babase is built upon standards[10] and popular, widely deployed, Open Source and Free Software. This means, among other things, that the tools used to build and run Babase are very likely available to anyone free of cost, and that the skill-sets required for the system maintenance of and, to some extent, use of Babase are readily learned[11] and unlikely to become obsolete[12].[13] The Babase source code itself is Free Software[14] and may be downloaded by the public.[15]
Note
The database design attempts 5th normal form, no redundant data, no empty data elements allowed, etc. What we've actually wound up with is about 3rd normal form.
The Babase system is accessed over the web. Any web browser may be used to view the data using the phpPgAdmin generic database interface. More advanced usage of the website will likely require a web browser that conforms to the international standards for the web defined by the World Wide Web Consortium , otherwise known as the W3C ,as we have put forth no particular effort to accommodate non-standards conforming browsers. The browser must support CSS2 style sheets and XHTML 1.0. Note that at the time of this writing Microsoft Internet Explorer does not provide adequate style sheet support. Other browsers that do have such support include Mozilla , Mozilla Firefox ,Apple's Safari ,and Opera. The W3.org site maintains a list of browsers supporting style sheets.
Babase's URL (web address)
is https://papio.biology.duke.edu/ . Be sure to
type the s in https . This secures
your web connection.
You must access most of the Babase web site using a secure communications protocol ( HTTPS ) that encrypts all communication to foil eavesdroppers and checks the identity of the web site itself. The Babase project has signed its own security certificate, the certificate that ensures you are talking with the website you think you are.[16] Our certificate expires annually and is re-generated.
Your browser probably will not trust that our website is who it says it is and so will very likely object when you first access the Babase web site, and annually thereafter. You may tell your browser to accept our certificate permanently.
Resources related to Babase include:
The Babase Pocket Reference is available as a PDF (4.8MB) and as a web page.
Babase Wiki, wherein lives much goodness including tutorial's, guides, and documentation on various data and material related to but not yet part of Babase (like the tables in the babase_pending schema)
The Babase Web site
The Amboseli Baboon Research Project's web site
The Protocol for Data Management: Amboseli Baboon Project
The Babase mailing list and its associated archive
The PostgreSQL web site, documentation, PostgreSQL's mailing list for beginner's questions, PostgreSQL's mailing list for SQL questions, and the general mailing list
The phpPgAdmin web site, documentation, and help forum
Some vagaries of spreadsheets, Excel in particular, that can affect their use as an source of Babase data are found in the page Spreadsheet Addiction, although the main focus of the page is the appropriate and inappropriate uses of spreadsheets in comparison with alternatives.
Babase users are encouraged to ask questions, both on the Babase mailing list and on the mailing lists setup for questions on the software that Babase is made of.
[1] Immediate reporting of some unusual conditions could be added to Babase at a later date.
[2] More or less. The last actual SQL standard was issued a very long time ago. None the less SQL is pervasive and, although specific SQL statements may not always be, the skill set involved in SQL use is quite portable.
[3] There are many PostgreSQL user interfaces available, although at the time of this writing only 2, phpPgAdmin and psql, are installed on the Babase database server. Many of these front-ends must be installed on the local workstation. These may require that the Babase VPN be running before initiating a connection to the database. Some of available front-ends may be found via the PostgreSQL FAQ question regarding graphical user interfaces for PostgreSQL.
[4] It's those pesky unpredictable users. Computer software would be a lot easier to write if it weren't for users always messing things up and then insisting on knowing what happened.
[5] A process which, admittedly, sometimes conflicts with the notion of easily maintaining the software. On the other hand when done right this approach does wonders for data integrity.
[7] These features also free the user from “software interface lock-in”. The database may be accessed and maintained with the software of choice. Data integrity, in both raw and derived data, is assured. Significantly, these features are those that allow Babase to leverage generic programs, using them for the bulk of its user interface as opposed to building a custom, Babase specific, interface.
[8] Of course, because Babase has no designated front-end and so much data validation takes place inside the database itself, any program able to talk with PostgreSQL, the database engine Babase uses, can be used to import data into the database. So there are no real limits on how data must be structured for import into Babase.
[9] There are 2 Unix shell programs that provide peripheral utility; both do tasks that can be done with other tools but are handy to have automated. The use of these programs are documented on the Babase Wiki. Comprehensive documentation of these programs should probably be added to this document.
The Unix Shell Programs
babase-copy-babase-schemaCopies the entire content of the babase schema from one database to another.
babase-user-addAdds a postgresql user, granting the permission to use Babase
There is also the ranker program, which runs on the local workstation and uses the Internet to communicate with the database. Developed separately from the rest of Babase, neither the source code management of nor the documentation for the ranker program is particularly well integrated into Babase.
[10] Actual standards, not de facto ones.
[11] Because open standards and the documentation for Open Source and Free Software programs are available, without cost; and because the inherently transparent and public nature of open standards, Open Source and Free Software leads not only to a wealth of good instructional material freely available on the Internet but also rounds out the basic requirements of a complete learning environment by ensuring that the software itself is available to everyone.
[12] Because once software is released and distributed under a Free or Open Source license it cannot be locked away and made unavailable, and because open standards are rarely changed in a backwards-incompatible way.
[13] Consequently the skills are rather widely available. The difficult part, as always, is finding the all of the relevant skills at once. For more on this see The Babase Program Code section.
[14] Presently licensed under the GPL Version 3 or later.
[15] Babase database content is not available to the public.
[16] We do this rather than paying one of the regular certification authorities to validate our identity. These certification authorities appear to validate the identity of their customers by virtue of little more than having successfully been paid.
Table of Contents
Databases are collections of information, all of which can be queried and otherwise manipulated alone or in aggregation with all other database content.[17] Babase contains three databases.
The babase database contains the “real” information. All research takes place in this database.
The babase_copy database contains a copy of the babase database. It is a place to try out dangerous things that might break the babase database.
Each user is given a login and a password they must use to gain access to the database. It is good form to change your password occasionally.[18]
The database can grant specific users various levels of access to specific tables, although such access is not common as it is difficult to administer and maintain such a fine grained degree of control. For further information see the PostgreSQL documentation on Database Users and Privileges.
Rather than maintain database access privileges on a per-user basis it is more convenient to place users in groups and then grant these groups different levels of database access.
Babase contains the following groups:
The members of this group have read access to Babase data and cannot add, delete, or otherwise alter any of the data.
The members of this group have unlimited rights to the Babase data. They may add data, delete data, or alter existing data. They may not, however, alter the structure of the babase database or change the rules to which the data are required to conform. Thus, they may not add or delete tables, alter triggers, or write or replace stored procedures.
Schemas partition databases. Tables, procedures, triggers, and so forth are all kept in schemas. Schemas are like sub-databases within a database. The salient difference between schemas and databases is that a single SQL statement can refer to objects in the different schemas of the parent database, but cannot refer to objects in other databases -- tables within a database can be related, but tables in different databases cannot. Babase uses schemas to partition each database into areas where users have a greater or lesser degree of freedom to make changes. For further information on schemas see the schema documentation for PostgreSQL.
Each database is divided into the same schemas. That is, each schema described below exists within each of the databases described here.
The system looks at the different schemas for objects, for example table names appearing in SQL queries, in the order in which the schemas are listed below. If the table does not appear in the first schema it looks in the second, and so forth. As soon as a table is found with the name given, that table is used and the search stops.
To explicitly reference an object in a specific schema, place the name of the schema in front of the object, separating the two with a period (e.g. schemaname.tablename).
The babase schema holds the “official” Babase tables. Everything in the babase schema is documented and supported.
In this schema the babase_readers and babase_editors have the access described above.
Babase contains a number of schemas that exist to simplify things for those interested only in particular portions of Babase. These schemas contain nothing but views that reference other parts of Babase, the parts that are especially relevant and useful to those interested only in one of the broad categories of Babase data. These schemas and their corresponding categories are:
| Schema | Category |
|---|---|
babase_cycles_views | Sexual Cycles |
babase_darting_views | Darting |
babase_demog_views | Group Membership and Life Events |
babase_physical_traits_views | Physical Traits |
babase_social_views | Social and Multiparty Interactions |
babase_support_views | Support Tables |
babase_swerb_views | SWERB Data (Group-level Geolocation Data) |
babase_weather_views | Weather Data |
babase_group_views | Views Which Add Gid To Tables |
These schemas provide an overview of the major areas of Babase. They should be especially useful to those starting out with Babase or those interested only in particular portions of Babase data.
The views in these schemas may only be queried. Any
updating of Babase data must be done in the
babase schema.
Note
Some of Babase's tables and views appear in more than one of these schemas, some in none.
Warning
Do not create any views that reference the views in
these schemas. Reference the babase schema
instead. Any views created that reference anything in these
“category schemas” will be destroyed on
occasion as Babase is modified.
The babase_history schema contains a table for each temporal table in the babase schema. The tables in this schema store the "old" versions of data from those temporal tables, allowing the ability to query for earlier versions of the data. See the Temporal Tables and babase_history appendix for more details.
The name of each table in this schema should be a concatenation of 1) the name of the related babase schema table, and 2) "_HISTORY". For example, a table in the babase schema called SOMETABLE would have a table in the babase_history schema called SOMETABLE_HISTORY.
Members of the babase_readers and babase_editors
groups both have the same permissions in the babase_history
schema: they have read access to the data but cannot perform
INSERT, UPDATE, or
DELETE commands in any tables[19], nor can they add new tables to the schema.
Only administrators are allowed to perform these
actions.
The babase_pending schema holds tables pending planned integration into Babase. The tables in this schema are intended to be used with the “official” Babase tables but, unlike the “official” Babase tables, there is no automated validation process and the table structure has not been thoroughly reviewed. The tables in babase_pending are to be used but their content and structure may change when officially incorporated into Babase.
Documentation on the content of the babase_pending schema may be found on the babase_pending page of the Babase Wiki.
The difference between this schema and the sandbox schema is in the permissions granted.
Members of the babase_readers group have the same permissions they do in the babase schema, they have read access to the data but cannot add, delete or modify it. However, unlike in the babase schema, individual users may be granted the right to add, delete, or change data on a table-by-table basis.
The sandbox schema holds tables that are used together with the “official” Babase tables but have not yet made it into the Babase project. They will not be documented in the Babase documentation.
The groups have the following permissions:
The babase_readers have all the permissions in the sandbox schema that the babase_editors have in the babase schema. They may add, delete, or modify any information in the schema but may not alter the structure of the schema by adding or removing tables, procedures, triggers, or anything else.
The babase_editors have all the permissions of the babase_readers, plus they may add or delete tables, stored procedures, or any other sort of object necessary to control the structure of the data.
Because of the schema search order the schema name must be used to qualify anything created in the sandbox schema. E.g.
Example 2.1. Creating table foo in the sandbox
schema
CREATE TABLE sandbox.foo (somecolumn INTEGER);
PostgreSQL, the database underlying Babase, is secure by default. This means that any tables or other database objects cannot be accessed by anyone but their creator without permission of the creator. Babase_editors who create tables in the sandbox schema should use the GRANT statement to grant access to Babase's other users.
This is done as follows:
Example 2.2. Granting permission to table
foo in the sandbox schema
GRANT ALL ON sandbox.foo TO GROUP babase_editors;
GRANT SELECT ON sandbox.foo TO GROUP babase_readers;
There is one other issue. Only the creator of a table can change its structure -- to add another column, change the table name, etc. And only the creator can destroy (DROP) the table.
The devel schema holds tables undergoing integration into Babase. Normally it is empty, but during the design and development of new tables it may contain the tables being developed.
The tables in this schema do not necessarily contain valid or finalized data and so are not expected to be used for other than developmental purposes.
Permissions are granted in the devel schema on the same basis as the granting of permissions in the babase schema.
The difference between this schema and the sandbox schema is that the development tools support the creation and modification of the tables in the devel schema, which facilitates the movement of tables from the devel schema into the babase schema.
Each user has her own schema, a schema named with the user's login. Users have permissions to do anything they want in their own schemas, and no permissions whatsoever to anybody else's schema. A user's schema is private.
Caution
Users are not encouraged to grant others permissions to the tables in their schema, as shown in the Section: “The sandbox schema” above. A user's schema is deleted when she leaves Babase. All shared tables belong in the sandbox schema where they can be maintained without regard to personnel changes.
Because of the schema search order the schema name must be used to qualify anything created in the user's schema. E.g.
Example 2.3. Creating table foo in user
mylogin's schema
CREATE TABLE mylogin.foo (somecolumn INTEGER);
The data in Babase are stored in tables. Tables can be visualized as grids, with rows and columns. Each row represents a single real-world thing or event, an entity, e.g. a baboon. Each cell in the row contains a single unit of information, e.g. a birth date, a name, and a sex. The row holds the entirety of the information belonging to the entity as an isolated thing, e.g. baboon database entities consist of a birth date, a name, and a sex. Each column contains one and only one kind of information, e.g. birth date.
Table 2.1 is an example of a database table that might be used to represent baboons, one baboon per row. Notice that each cell contains one and exactly one unit of information.
Table 2.1. A Simple Database Table
| Birth | Name | Sex |
|---|---|---|
| May 23, 1707 | Alice | Female |
| February 12, 1809 | Bob | Male |
| July 22, 1822 | Carol | Female |
Anyone working with Babase will require a familiarity with the database's tables. An understanding of the entity each row represents is critical when working with a table. The tables below provide short definitions of the entities each The babase schema table holds in its rows.
Some of the tables in Babase exist to define a vocabulary. These are the support tables. For lack of a better term, the remainder of the tables are labeled “main tables” in Table 2.2.
Warning
Tables which have names ending in “_DATA” should not be used, there is always a view of the data in these tables that may be used in their place. Tables ending in “_DATA” may change in future Babase minor releases, breaking queries and programs which use the table. Use of the corresponding views will ensure compatibility with future Babase releases.
Table 2.2. The Main Babase Tables
| Group Membership and Life Events | |
| Table | One row for each |
| ALTERNATE_SNAMES | rescinded sname |
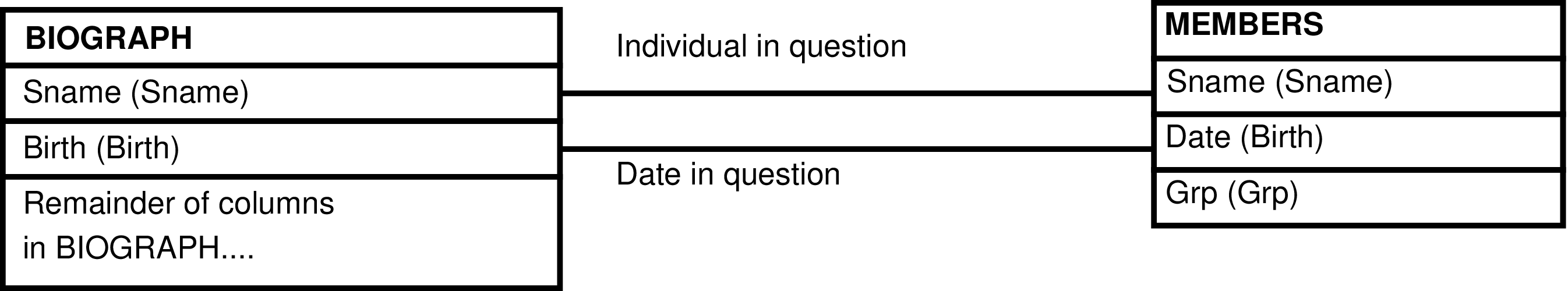
| BIOGRAPH | animal, including fetuses |
| CENSUS | day each individual is (or is not) observed in a group |
| CONSORTDATES | male who has a known first consortship |
| DEMOG | mention of an individual's presence in a group within a field textual note |
| DISPERSEDATES | male who has left his maternal study group |
| GROUPS | group (including solitary males) |
| MATUREDATES | individual who is sexually mature |
| RANKDATES | individual[a] who has attained adult rank |
| Analyzed: Group Membership and Life Events | |
| Table | One row for each |
| DADS_ANALYSES | paternity analysis |
| DADS_CONSENSUS | kid with a known dad |
| DADS_EVIDENCE | datum used in a paternity analysis |
| MEMBERS | day each individual is alive |
| RANKS | month each individual is ranked in each group |
| RESIDENCIES | bout of each individual's residency |
| Physical Traits | |
| Table | One row for each |
| WP_AFFECTEDPARTS | body part affected by a specific wound/pathology |
| WP_DETAILS | wound or pathology cluster indicated on a report |
| WP_HEALUPDATES | update on progress of wound/pathology healing |
| WP_REPORTS | wound/pathology report |
| Analyzed: Physical Traits | |
| Table | One row for each |
| HORMONE_KITS | kit or protocol used to assay hormone concentration |
| HORMONE_PREP_DATA | laboratory preparation performed on a sample in the specified series |
| HORMONE_PREP_SERIES | series of preparations and assays performed on a sample |
| HORMONE_RESULT_DATA | assay for hormone concentration in a sample |
| HORMONE_SAMPLE_DATA | tissue sample used in hormone analysis |
| HYBRIDGENE_ANALYSES | analysis of genetic hybrid scores |
| HYBRIDGENE_SCORES | genetic hybrid score for an individual from an analysis |
| HYBRIDMORPH_OBSERVERS | observer in a morphological hybrid score report |
| HYBRIDMORPH_REPORTS | morphological hybrid scoring event, per scored individual |
| HYBRIDMORPH_SCORE_DATA | morphological hybrid score for a particular trait |
| Sexual Cycles | |
| Table | One row for each |
| CYCGAPS | female for each initiation or cessation of a continuous period of observation |
| CYCLES | female's cycle (complete or not) |
| CYCPOINTS | Mdate (menses), Tdate (turgesence onset), or Ddate (deturgesence onset) date of each female |
| PREGS | time a female becomes pregnant |
| SEXSKINS | sexskin measurement of each female |
| The Sexual Cycle Day-By-Day Tables | |
| Table | One row for each |
| CYCGAPDAYS | female for each day within a period during which there is not continuous observation |
| CYCSTATS | day each female is cycling -- by M, T and Ddates |
| MDINTERVALS | day each female is cycling and is between M and Ddates |
| MMINTERVALS | day each female is cycling -- by Mdates |
| REPSTATS | day each female has a known reproductive state |
| Social and Multiparty Interactions | |
| Table | One row for each |
| ALLMISCS | “free form” all-occurrences datum |
| CONSORTS | multiparty dispute over a consortship |
| FPOINTS | point observation of a mature female |
| INTERACT_DATA | interaction between individuals |
| MPIS | collection of multiparty interactions |
| MPI_DATA | single dyadic interaction of a multiparty interaction collection |
| MPI_PARTS | participant in a dyadic interaction of a multiparty interaction collection |
| PARTS | participant in each interaction |
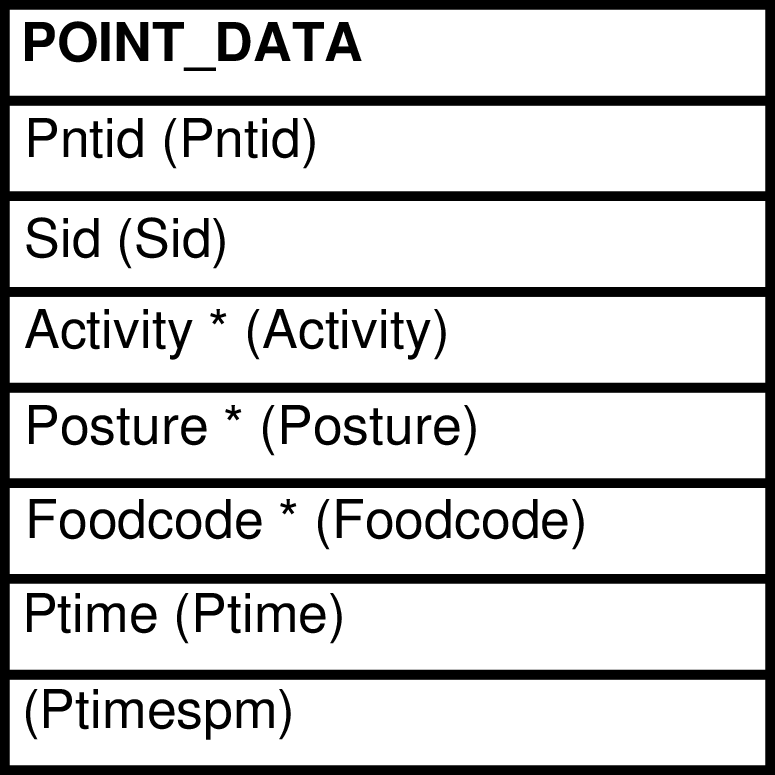
| POINT_DATA | individual point observation |
| NEIGHBORS | neighbor recorded in each point sample |
| SAMPLES | focal sample |
| Darting | |
| Table | One row for each |
| ANESTHS | time additional sedation is administered to a darted individual |
| BODYTEMPS | body temperature measurement taken of a darted individual |
| CHESTS | chest circumference measurement made of a darted individual |
| CROWNRUMPS | crown to rump measurement made of a darted individual |
| DART_SAMPLES | sample type collected at each darting |
| DARTINGS | darting of an animal when data was collected |
| DPHYS | darting event during which physiological measurements were taken |
| HUMERUSES | humerous length measurement made of a darted individual |
| PCVS | packed cell volume measurement taken from a darted individual |
| TEETH | possible tooth site within the mouth on which data was collected for every darting event during which dentition data was collected |
| TESTES_ARC | every testicle width/length measurement recorded, as measured along a portion of the circumference |
| TESTES_DIAM | every testicle width/length measurement recorded, as measured along the diameter |
| TICKS | darting event during which data on ticks and other parasites were recorded |
| ULNAS | ulna length measurement made of a darted individual |
| VAGINAL_PHS | vaginal pH measurement made of a darted individual |
| Analyzed: Darting | |
| Table | One row for each |
| FLOW_CYTOMETRY | flow cytometric analysis of a blood sample collected during a darting |
| WBC_COUNTS | count from a blood smear collected during a darting |
| Inventory | |
| Table | One row for each |
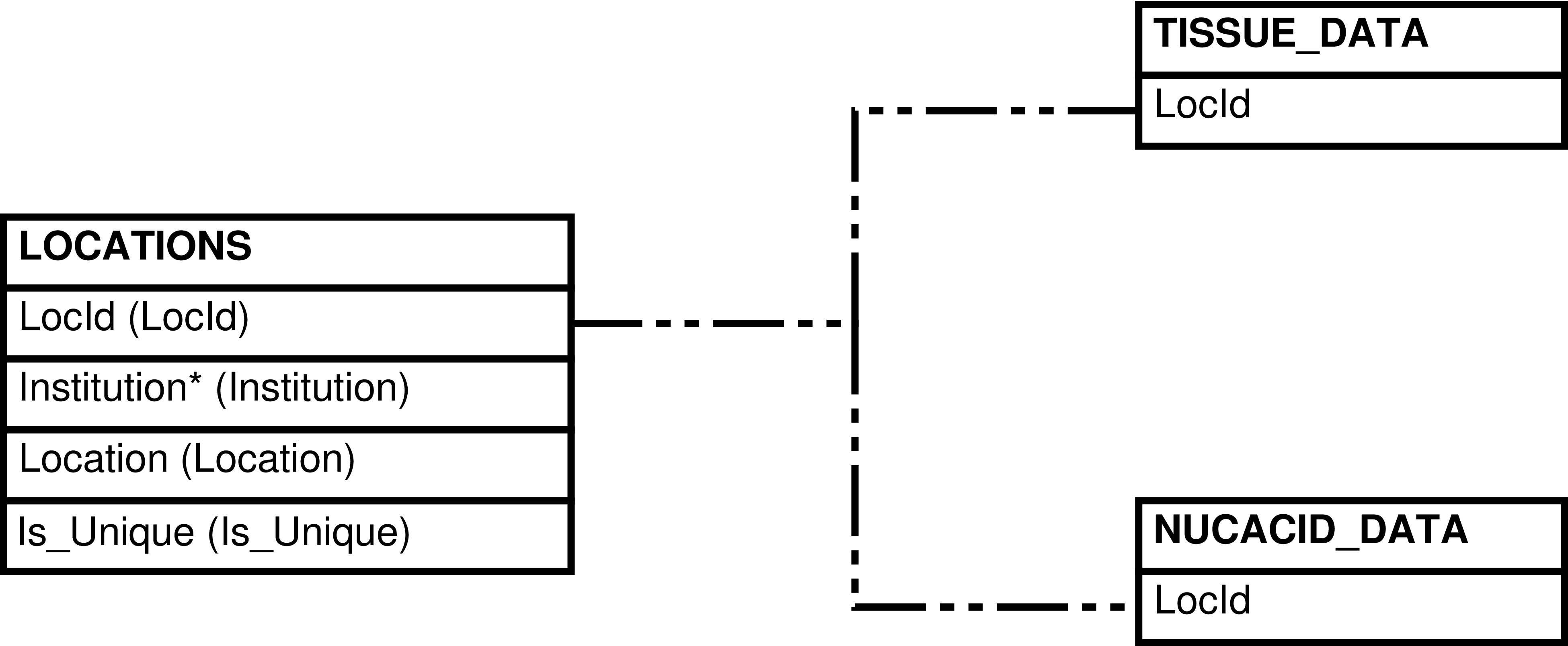
| LOCATIONS | Location that can be used to store tissue and nucleic acid samples |
| NUCACID_CONC_DATA | Quantification of a nucleic acid sample's concentration |
| NUCACID_DATA | Nucleic acid sample that is or ever has been in the inventory |
| NUCACID_LOCAL_IDS | Name/ID used to identify a nucleic acid sample at a particular institution |
| NUCACID_SOURCES | Nucleic acid sample that has another nucleic acid sample as its source |
| POPULATIONS | Study population under observation or from which tissue or nucleic acid samples have been collected |
| TISSUE_DATA | Tissue sample that is or ever has been in the inventory |
| TISSUE_LOCAL_IDS | Name/ID used to identify a tissue sample at a particular institution |
| TISSUE_SOURCES | Tissue sample that has another tissue sample as its source |
| UNIQUE_INDIVS | Individual under observation or from whom tissue or nucleic acid samples have been collected |
| SWERB Data (Group-level Geolocation Data) | |
| Table | One row for each |
| AERIALS | aerial photo used for map quadrant specification |
| GPS_UNITS | GPS device |
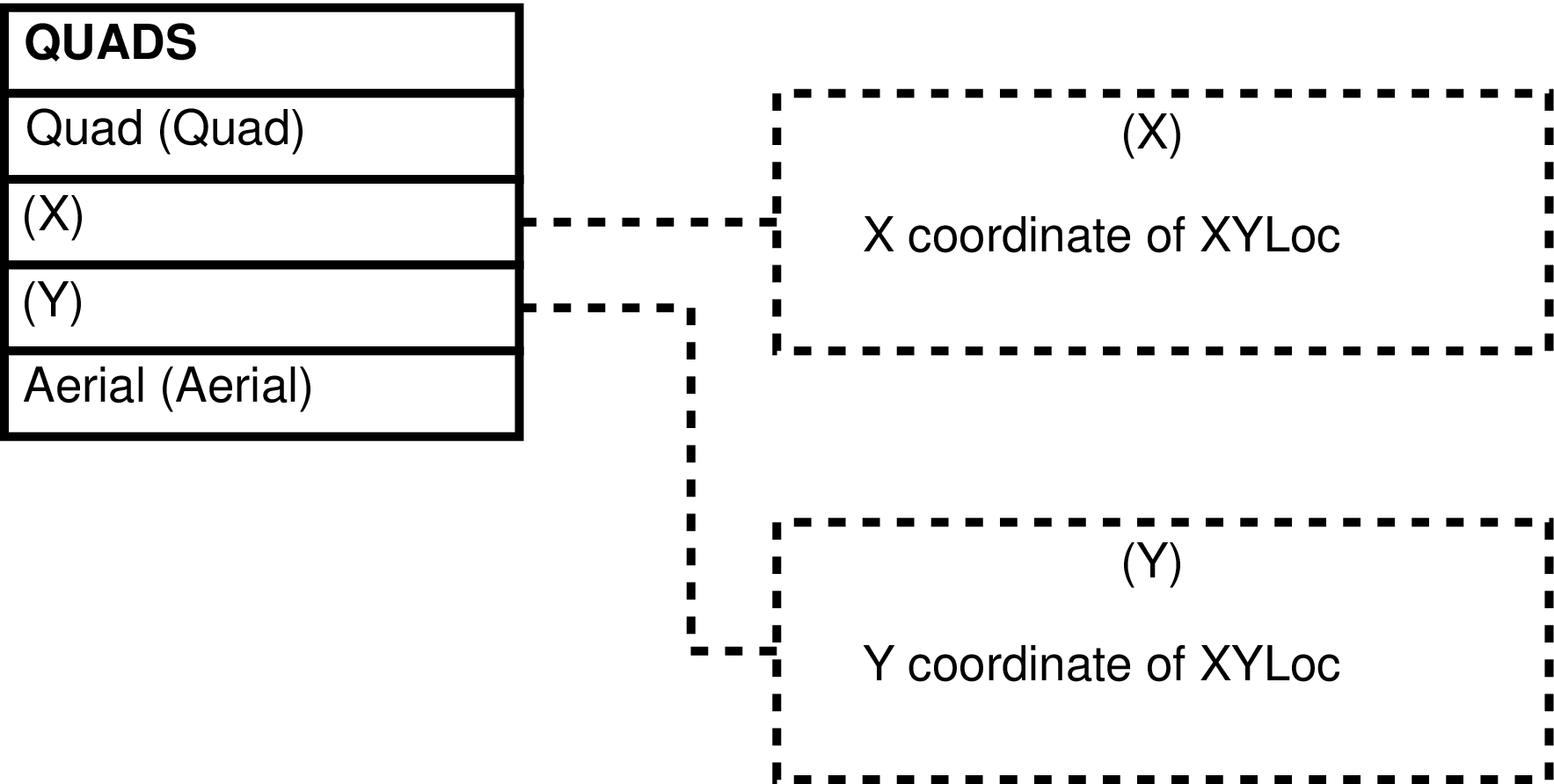
| QUAD_DATA | SWERB map quadrant |
| SWERB_BES | uninterrupted bout of group-level observation |
| SWERB_DATA | event related to group-level geolocation |
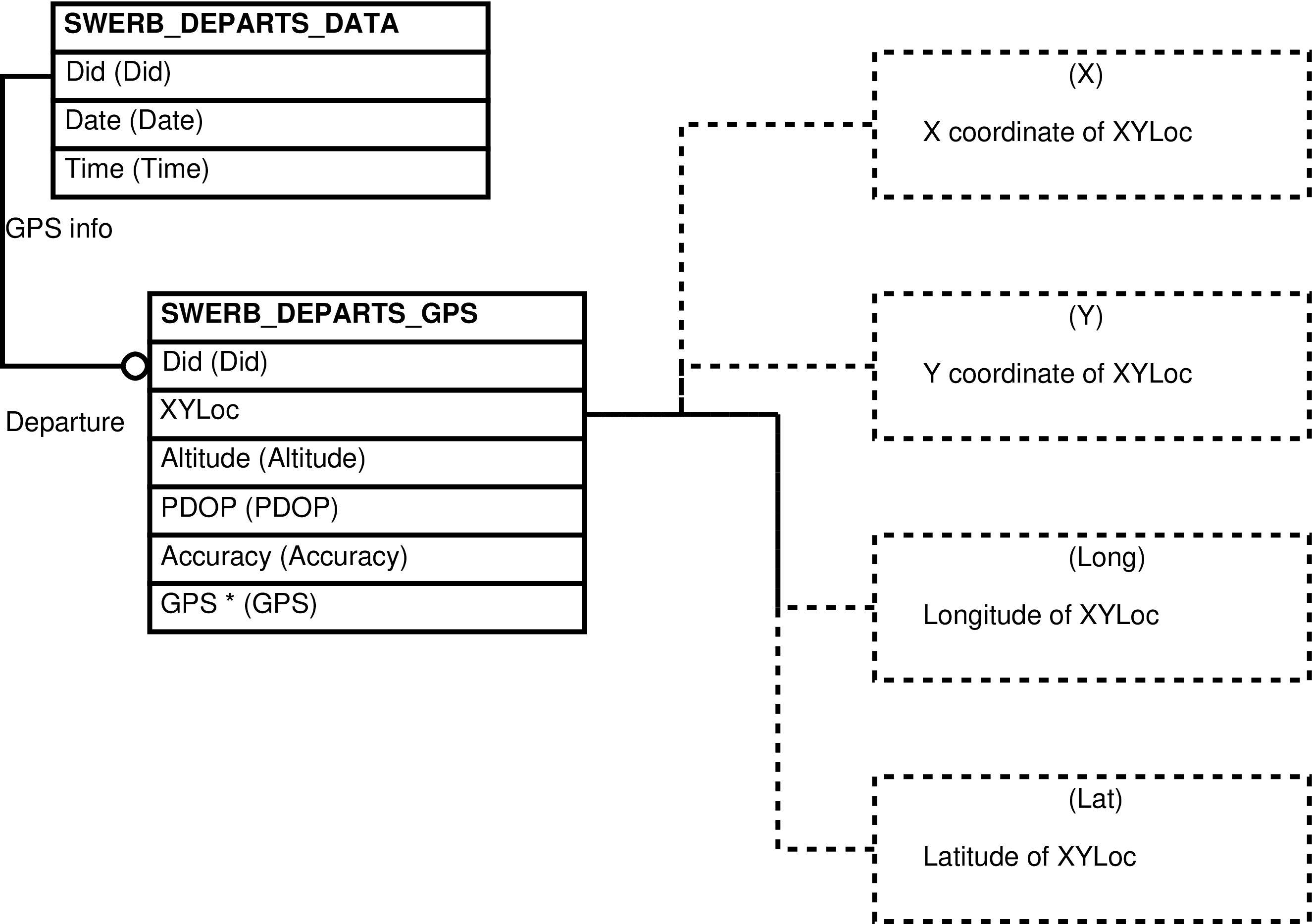
| SWERB_DEPARTS_DATA | departure from camp of a observation team which collected SWERB data |
| SWERB_GWS | geolocated physical object (grove or waterhole) |
| SWERB_GW_LOC_DATA | recorded location of a geolocated physical object (grove or waterhole) |
| SWERB_LOC_DATA | observation of a group at a time at a geolocated physical object |
| SWERB_LOC_DATA_CONFIDENCES | analyzed observation of a location |
| SWERB_LOC_GPS | observation of a group at a time at a geolocated physical object made using gps units and a protocol that requires 2 waypoint readings |
| SWERB_OBSERVERS | departure from camp of an observer who drove or collected SWERB data |
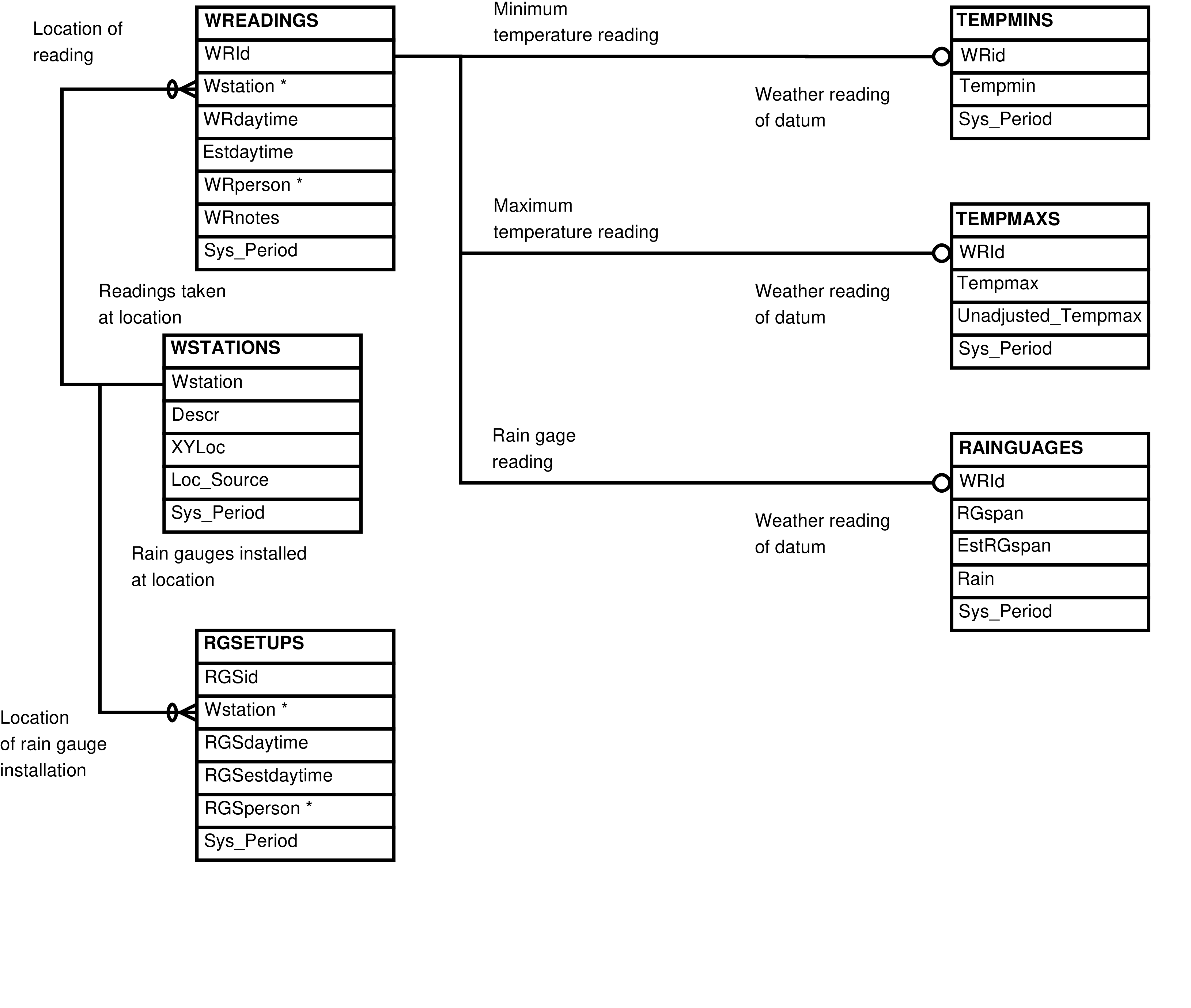
| Weather Data | |
| Table | One row for each |
| RAINGAUGES | rain gauge reading |
| RGSETUPS | rain gauge installation |
| TEMPMAXS | maximum temperature reading |
| TEMPMINS | minimum temperature reading |
| DIGITAL_WEATHER | digital weather reading reported from an electronic weather collection device |
| WREADINGS | manually collected meteorological data collection event |
| Metadata | |
| Table | One row for each |
| BEHAVE_GAPS | gap in observation |
The significant aspects of the the support tables are: the Id column -- the name of the column holding the vocabulary term, which columns of which tables use the vocabulary, and what sort of vocabulary the table defines. Table 2.3 summarizes this information.
Note
The Id columns throughout Babase do not allow values
that are NULL, or which are textual but contain no
characters, or which consist solely of spaces.
Table 2.3. The Babase Support Tables
| General Support Tables | |||
| Table | Id Column | Related Column(s) | One entry for every possible choice of... |
| BODYPARTS | Bodypart | TICKS.Bodypart, BODYPARTS.Bodyregion, WP_AFFECTEDPARTS.Bodypart | part of the body |
| LAB_PERSONNEL | Initials | HYBRIDGENE_ANALYSES.Analyzed_By, NUCACID_CREATORS.Creator, WBC_COUNTS.Counted_By | person who generates data, usually in a lab setting |
| OBSERVERS | Initials | SAMPLES.Observer, WREADINGS.WRperson, RGSETUPS.RGSPerson, CROWNRUMPS.CRobserver, CHESTS.Chobserver, ULNAS.Ulobserver, HUMERUSES.Huobserver, SWERB_OBSERVERS.Observer | person who record observational data |
| OBSERVER_ROLES | Initials | OBSERVERS.Role, OBSERVERS.SWERB_Observer_Role, OBSERVERS.SWERB_Driver_Role, SWERB_OBSERVERS.Role | way in which a person can be involved in the data collection process |
| UNKSNAMES | Unksname | NEIGHBORS.Unksname and the SWERB_UPLOAD view | problem in identifying neighbor of focal during point sampling or in identifying a lone male in a SWERB other group observation |
| Group Membership and Life Events | |||
| Table | Id Column | Related Column(s) | One entry for every possible choice of... |
| BSTATUSES | Bstatus | BIOGRAPH.Bstatus | birthday estimation accuracy |
| CONFIDENCES | Confidence | BIOGRAPH.DcauseNatureConfidence, BIOGRAPH.DcauseAgentConfidence, DISPERSEDATES.Dispconfidence, BIOGRAPH.Matgrpconfidence | degree of certitude in nature of death, agent of death, disperse date assignment, or maternal group assignment |
| DADS_COMPLETENESS | Completeness | DADS_CONSENSUS.Completeness | category of analysis completeness |
| DADS_EVIDENCE_TERMS | Term | DADS_EVIDENCE.Term | kind of evidence used in a paternity analysis |
| DADS_MISMATCHES | Mismatch | DADS_CONSENSUS.Mismatch | category of analysis completeness |
| DCAUSES | Dcause | BIOGRAPH.Dcause | cause of death |
| DEATHNATURES | Nature | DCAUSES.Nature | reason for death |
| DEMOG_REFERENCES | Reference | DEMOG.Reference | data source for demography notes |
| MSTATUSES | Mstatus | MATUREDATES.Matured, RANKDATES.Ranked | maturity marker date estimation process |
| RNKTYPES | Rnktype | RANKS.Rnktype | rank ordering assigned to subject and month |
| STATUSES | Status | BIOGRAPH.Status | baboon alive at last observation |
| Physical Traits | |||
| Table | Id Column | Related Column(s) | One entry for every possible choice of... |
| HORMONE_IDS | Hormone | HORMONE_KITS.Hormone | hormone that may be extracted and assayed for |
| HORMONE_PREP_PROCEDURES | Id | HORMONE_PREP_DATA.Procedure | procedure that may be performed in preparation for a hormone assay |
| HYBRIDGENE_SOFTWARE | Software | HYBRIDGENE_ANALYSES.Software | software used for genetic hybrid score analysis |
| MARKERS | Marker | HYBRIDGENE_ANALYSES.Marker | type of genetic marker used for genetic hybrid score analysis |
| WP_HEALSTATUSES | Healstatus | WP_HEALUPDATES.HealStatus | healing progress used in healing updates |
| WP_REPORTSTATES | ReportState | WP_REPORTS.ReportState | status of wound/pathology report |
| WP_WOUNDPATHCODES | WoundPathCode | WP_DETAILS.WoundPathCode | wound or pathology |
| Social and Multiparty Interactions | |||
| Table | Id Column | Related Column(s) | One entry for every possible choice of... |
| ACTIVITIES | Activity | POINT_DATA.Activity | activity classification |
| ACTS | Act | INTERACT_DATA.Act | interaction classification |
| DATA_STRUCTURES | Data_Structure | SETUPIDS.Data_Structure | version of data structure produced by the data collection devices |
| CONTEXT_TYPES | Context_type | MPIS.Context_type | context in which a multiparty interaction occurs |
| FOODCODES | Foodcode | POINT_DATA.Foodcode | name of a food item |
| FOODTYPES | Ftype | FOODCODES.Ftype | food category |
| KIDCONTACTS | Kidcontact | FPOINTS.Kidcontact | spatial relationship between mother and infant |
| MPIACTS | Mpiact | MPI_DATA.MPIAct | multiparty interaction classification |
| NCODES | Ncode | NEIGHBORS.Ncode | neighbor classification |
| PARTUNKS | Unksname | MPI_PARTS.Unksname | problem in identifying participant in a multiparty interaction |
| POSTURES | Posture | POINT_DATA.Posture | designated posture |
| PROGRAMIDS | Programid | SAMPLES.Programid | version of each program used on the devices to collect focal sampling data |
| SAMPLES_COLLECTION_SYSTEMS | Collection_System | SAMPLES.Collection_System | device or "system" used in the field for collecting focal sampling data |
| SETUPIDS | Setupid | SAMPLES.Setupid | setupfile used on the devices to collect focal sampling data |
| STYPES | SType | SAMPLES.SType | protocol for focal sampling data collection |
| STYPES_ACTIVITIES | SType-Activity pair | SAMPLES.SType, ACTIVITIES.Activity | activity classification allowed to be used in each focal sampling protocol |
| STYPES_NCODES | SType-Ncode pair | SAMPLES.SType, NCODES.Ncode | neighbor classification allowed to be used in each focal sampling protocol |
| STYPES_POSTURES | SType-Posture pair | SAMPLES.SType, POSTURES.Posture | posture classification allowed to be used in each focal sampling protocol |
| SUCKLES | Suckle | FPOINTS.Kidsuckle | infant suckling activity |
| Sexual Cycles and The Sexual Cycle Day-By-Day Tables | |||
| Table | Id Column | Related Column(s) | One entry for every possible choice of... |
| PCSCOLORS | Color | SEXSKINS.Color | paracallosal skin coloration |
| Darting | |||
| Table | Id Column | Related Column(s) | One entry for every possible choice of... |
| DART_SAMPLE_CATS | Ds_cat | DART_SAMPLE_CATS.DS_Cat | category of darting sample type |
| DART_SAMPLE_TYPES | DS_Type | DART_SAMPLE_TYPES.DS_Type | type of sample collected during dartings |
| DRUGS | Drug | DRUGS.Drug | anesthetic drug |
| LYMPHSTATES | Lymphstate | DPHYS.Ringnode, DPHYS.Lingnode, DPHYS.Raxnode, DPHYS.Laxnode, DPHYS.Lsubmandnode, DPHYS.Rsubmandnode | lymph node condition |
| PARASITES | PARASITE | TICKS.Tickkind | parasite species, species developmental stage, or kind of parasite sign counted |
| TCONDITIONS | Tcondition | TEETH.Tcondition | physical condition of a tooth |
| TICKSTATUSES | Tickstatus | TICKS.Tickstatus | parasite count outcome category |
| TOOTHCODES | Tooth | TEETH.Tooth | adult or deciduous tooth |
| TOOTHSITES | Toothsite | TOOTHCODES.Toothsite | dental site within the mouth |
| TSTATES | Tstate | TEETH.Tstate | tooth “presence” |
| Inventory | |||
| Table | Id Column | Related Column(s) | One entry for every possible ... |
| INSTITUTIONS | Institution | LOCATIONS.Institution, NUCACID_LOCAL_IDS.Institution, TISSUE_LOCAL_IDS.Institution | possible locale where tissue and nucleic acid samples can be stored or used |
| MISID_STATUSES | Misid_Status | TISSUE_DATA.Misid_Status | level of confidence in the identity of a tissue sample |
| NUCACID_CONC_METHODS | Conc_Method | NUCACID_CONC_DATA.Conc_Method | method used for quantifying nucleic acid concentrations |
| NUCACID_CREATION_METHODS | Creation_Method | NUCACID_DATA.Creation_Method | method used for creating nucleic acid samples |
| NUCACID_TYPES | NucAcid_Type | NUCACID_DATA.NucAcid_Type | type of nucleic acid sample |
| STORAGE_MEDIA | Storage_Medium | TISSUE_DATA.Storage_Medium | medium used for storage/archiving of tissue samples |
| TISSUE_TYPES | Tissue_Type | TISSUE_DATA.Tissue_Type | type of tissue sample |
| SWERB Data (Group-level Geolocation Data) | |||
| Table | Id Column | Related Column(s) | One entry for every possible ... |
| ADCODES | ADCode | SWERB_LOC_DATA.ADcode | relationship between baboon groups and sleeping groves. |
| SWERB_LOC_CONFIDENCES | Conf | SWERB_LOC_DATA_CONFIDENCES.Confidence | confidence score used when analyzing the accuracy of a recorded observation of a location. |
| SWERB_LOC_STATUSES | Conf | SWERB_LOC_DATA.Loc_Status | status for a recorded observation of a location. |
| SWERB_TIME_SOURCES | Source | SWERB_BES.Bsource, SWERB_BES.Esource | data source used to estimate beginning and ending of observation bouts |
| SWERB_XYSOURCES (SWERB Time Sources) | Source | SWERB_GW_LOC_DATA.XYSource | data source used to obtain XY coordinates |
| Weather Data | |||
| Table | Id Column | Related Column(s) | One entry for every possible choice of... |
| WEATHER_SOFTWARES | WSoftware | DIGITAL_WEATHER.WSoftware | software used to retrieve data from an electronic weather collection instrument |
| WSTATIONS | Wstation | WREADINGS.Wstation | meteorological data collection location or device |
| Metadata | |||
| Table | Id Column | Related Column(s) | One entry for every possible choice of... |
| GAP_END_STATUSES | Gap_End_Status | BEHAVE_GAPS.Gap_End_Status | reason why an observation gap ended |
Beginning with Babase 5.0, nearly every table in Babase has a column called "Sys_Period", which shows the range of time when the data in a row is considered "valid". When a row in a table in the babase schema is updated or deleted, the "old" version is no longer "valid" and is saved in a corresponding table in the babase_history schema.
Note
All data in the babase schema are valid, simply by virtue of their being in that schema. Users should not let this discussion of validity mislead them into undue suspicion of the accuracy of the data.
Updates to this column should only be performed automatically by the system, when data are inserted, updated, or deleted. Manual updates to this column are only allowed when done by an admin[20].
In the babase
schema, the lower bound of the Sys_Period column indicates
when the row was last updated, when the row was inserted to
the table, or when the Sys_Period column was added to the
table, whichever is most recent. The upper bound of the
Sys_Period column for tables in that schema will always be
NULL, meaning "no end" (yet).
In the babase_history schema,
each row represents an old "version" of the row. In these
tables, the lower bound of the Sys_Period column is the
timestamp of the INSERT or
UPDATE that created that version of the
row, or the date and time that the Sys_Period column was added
to the original table, whichever is most recent. The upper
bound is the timestamp of the INSERT,
UPDATE, or DELETE that
rendered the row no longer "valid".
In all tables, this column is a timestamp range (with
time zone), with inclusive lower bound
and exclusive upper bound. The lower
bound cannot be NULL, and defaults to the
current_timestamp when the row is
inserted/updated.
Most tables have have an id, or key, column that contains
a number unique to that row within its table. The id can be
used, in perpetuity, to refer to its related row and
distinguish it from all the other rows of the table. Ids are
arbitrary, although for convenience they are often sequentially
generated integers. The name of the column is not always
Id, although it sometimes is.
A relationship is established between the rows of two tables when an id value from one table appears as data in the other. The relationship notion is made most clear by way of diagrams and examples. If the next paragraph is unclear, don't worry. Have a look at the Babase diagrams below by way of example and see if that does not clear things up. The relationship concept is at the heart of relational databases and, while the underlying idea is rather simple, it took many years to develop relational database concepts[21] so don't expect a full understanding immediately.
When an id value of a row in one table appears as data in a second table, the data in the second table can be used to retrieve the identified row from the first table.[22] When an id value of a row in the first table appears as data only once in the second table, the two tables are said to have a one-to-one relationship. One row in the first table relates to one (or possibly zero) row(s) in the second table. When a row's id value can appear in more than one row of a second table, the two tables are said to have a one-to-many relationship. One row of the first table can be related to many rows in the second table. One-to-many relationships are more common than one-to-one relationships. The relationship between the various Babase tables can be visualized in entity relationship diagrams, as shown here. In this diagram each table (entity) is a box, and each box contains a list of the table's columns. The lines between the boxes represent the relationships between the tables.
Note
If you have trouble viewing the diagrams in your browser, you may wish to view them in PDF format. The diagrams are available in The Babase Pocket Reference (approx. 4.8MB) in PDF form.
Views provide an alternative to direct reference of Babase tables. Views appear to be tables, but are really pre-composed queries into the underlying Babase tables. Views can be used almost anywhere in Babase in place of a table, specifically, they can be queried just like tables. An SQL query can freely intermix the use of tables and views.
Important
Babase uses views to hide implementation details, details that may change as Babase develops. Tables that have names ending in “_DATA” should not be used, there is always a view of the data in these tables that may be used in their place. Tables ending in “_DATA” may change in future Babase minor releases, breaking queries and programs that use the table. Use of the corresponding views will ensure compatibility with future Babase releases.
Views make it easy to reuse complex or commonly used queries, or portions of queries. They allow a database designed around the capabilities of the computer to be interacted with in a fashion that makes sense to people. Although the views do not appear in the entity relationship diagrams that document the underlying database, and so are omitted from the high level overview these diagrams provide, most Babase users will greatly benefit if they take the time to understand how the views fit into the overall database and will usually find it easier to work with the views than with the underlying tables.
Table 2.4. The Babase Views
| Group Membership and Life Events | |||
| View | One row for each | Purpose | Tables/Views used |
| CENSUS_DEMOG | CENSUS row | Maintenance of CENSUS rows that are extended with DEMOG information. | CENSUS, DEMOG |
| CENSUS_DEMOG_SORTED | CENSUS row | Maintenance of CENSUS_DEMOG rows in a pre-sorted fashion. | CENSUS, DEMOG |
| CYCPOINTS_CYCLES | CYCPOINTS row | Maintenance of CYCPOINTS rows that are extended with CYCLES information. | CYCLES, CYCPOINTS |
| CYCPOINTS_CYCLES_SORTED | CYCPOINTS row | The CYCPOINTS_CYCLES view sorted by CYCLES.Sname, by CYCPOINTS.Date. | CYCLES, CYCPOINTS |
| DEMOG_CENSUS | DEMOG row | Maintenance of DEMOG rows. | CENSUS, DEMOG |
| DEMOG_CENSUS_SORTED | CENSUS row | Maintenance of DEMOG_CENSUS rows in a pre-sorted fashion. | CENSUS, DEMOG |
| GROUPS_HISTORY | GROUPS row | Depiction of GROUPS rows in a more human-readable format. | GROUPS |
| PARENTS | BIOGRAPH
row for which there is either a row in MATERNITIES with a
record of the individual's mother or there is a row in DADS_CONSENSUS with a
record of the individual's father -- with a non-NULLDad_Consensus. | Easy access to parental information. | BIOGRAPH, MATERNITIES, DADS_CONSENSUS, MEMBERS |
| POTENTIAL_DADS | (completed) female reproductive event for every male more than 2192 days old (approximately 6 years) present in the mother's group during her fertile period | Research into paternity, especially the selection of potential fathers for further genetic testing. | MATERNITIES, MEMBERS (multiple times), ACTOR_ACTEES (multiple times), BIOGRAPH, RANKDATES, MATUREDATES |
| PROPORTIONAL_RANKS | RANKS row | Automatic calculation of proportional ranks from the ordinal ranks in RANKS. | RANKS |
| Physical Traits | |||
| View | One row for each | Purpose | Tables/Views used |
| ESTROGENS | HORMONE_RESULT_DATA row with an estrogen kit | Easy access to estrogen data. | BIOGRAPH, HORMONE_KITS, HORMONE_PREP_DATA, HORMONE_PREP_SERIES, HORMONE_RESULT_DATA, HORMONE_SAMPLE_DATA, TISSUE_DATA, UNIQUE_INDIVS |
| GLUCOCORTICOIDS | HORMONE_RESULT_DATA row with a glucocorticoid kit. | Easy access to glucocorticoid data. | BIOGRAPH, HORMONE_KITS, HORMONE_PREP_DATA, HORMONE_PREP_SERIES, HORMONE_RESULT_DATA, HORMONE_SAMPLE_DATA, TISSUE_DATA, UNIQUE_INDIVS |
| HORMONE_PREPS | HORMONE_PREP_DATA row | Presents HORMONE_PREP_DATA with identifying information from TISSUE_DATA and BIOGRAPH. Also useful for maintaining data in HORMONE_PREP_DATA. | BIOGRAPH, HORMONE_PREP_DATA, HORMONE_PREP_SERIES, HORMONE_SAMPLE_DATA, TISSUE_DATA, UNIQUE_INDIVS |
| HORMONE_RESULTS | HORMONE_RESULT_DATA row | Presents HORMONE_RESULT_DATA with identifying information from TISSUE_DATA and BIOGRAPH. Also useful for maintaining data in HORMONE_RESULT_DATA. | BIOGRAPH, HORMONE_KITS, HORMONE_PREP_SERIES, HORMONE_RESULT_DATA, HORMONE_SAMPLE_DATA, TISSUE_DATA, UNIQUE_INDIVS |
| HORMONE_SAMPLES | HORMONE_SAMPLE_DATA row | Presents HORMONE_SAMPLE_DATA with identifying information from TISSUE_DATA and BIOGRAPH. Also useful for maintaining data in HORMONE_SAMPLE_DATA. | BIOGRAPH, HORMONE_SAMPLE_DATA, TISSUE_DATA, UNIQUE_INDIVS |
| HYBRIDMORPH_SCORES | HYBRIDMORPH_REPORTS row | Viewing and uploading morphological hybrid score data. | HYBRIDMORPH_OBSERVERS, HYBRIDMORPH_REPORTS, HYBRIDMORPH_SCORE_DATA |
| PROGESTERONES | HORMONE_RESULT_DATA row with a progesterone kit. | Easy access to progesterone data. | BIOGRAPH, HORMONE_KITS, HORMONE_PREP_DATA, HORMONE_PREP_SERIES, HORMONE_RESULT_DATA, HORMONE_SAMPLE_DATA, TISSUE_DATA, UNIQUE_INDIVS |
| TESTOSTERONES | HORMONE_RESULT_DATA row with a testosterone kit. | Easy access to testosterone data. | BIOGRAPH, HORMONE_KITS, HORMONE_PREP_DATA, HORMONE_PREP_SERIES, HORMONE_RESULT_DATA, HORMONE_SAMPLE_DATA, TISSUE_DATA, UNIQUE_INDIVS |
| THYROID_HORMONES | HORMONE_RESULT_DATA row with a thyroid hormone kit. | Easy access to thyroid hormone data. | BIOGRAPH, HORMONE_KITS, HORMONE_PREP_DATA, HORMONE_PREP_SERIES, HORMONE_RESULT_DATA, HORMONE_SAMPLE_DATA, TISSUE_DATA, UNIQUE_INDIVS |
| WOUNDSPATHOLOGIES | WP_AFFECTEDPARTS row | Querying of wounds/pathologies data (without heal updates). | WP_REPORTS, WP_OBSERVERS, WP_DETAILS, WP_AFFECTEDPARTS, BODYPARTS |
| WP_DETAILS_AFFECTEDPARTS | WP_AFFECTEDPARTS row | Upload of WP_DETAILS and WP_AFFECTEDPARTS rows. | WP_DETAILS, WP_AFFECTEDPARTS, BODYPARTS |
| WP_HEALS | WP_HEALUPDATES row | Upload and viewing of WP_HEALUPDATES rows. | WP_REPORTS, WP_DETAILS, WP_AFFECTEDPARTS, BODYPARTS, WP_HEALUPDATES |
| WP_REPORTS_OBSERVERS | WP_REPORTS row | Upload of WP_REPORTS and WP_OBSERVERS rows. | WP_REPORTS, WP_OBSERVERS |
| Sexual Cycles | |||
| View | One row for each | Purpose | Tables/Views used |
| CYCLES_SEXSKINS | CYCLES row | Maintenance of SEXSKINS rows. | CYCLES, SEXSKINS |
| CYCLES_SEXSKINS_SORTED | CYCLES row | The CYCLES_SEXSKINS view sorted by CYCLES.Sname, by SEXSKINS.Date. | CYCLES, SEXSKINS |
| MATERNITIES | birth or fetal loss | Summarizes (completed) reproductive events. | BIOGRAPH, PREGS, CYCPOINTS, CYCLES |
| MTD_CYCLES | CYCLES row | Presents CYCLES together with Mdate, Tdate, and Ddate CYCPOINTS information for a view of an "entire" sexual cycle as a single row. | CYCLES, CYCPOINTS |
| SEXSKINS_CYCLES | SEXSKINS row | Maintenance of SEXSKINS rows. | CYCLES, SEXSKINS |
| SEXSKINS_CYCLES_SORTED | SEXSKINS row | The SEXSKINS_CYCLES view sorted by CYCLES.Sname, by SEXSKINS.Date. | CYCLES, SEXSKINS |
| SEXSKINS_REPRO_NOTES | SEXSKINS row, or REPRO_NOTES row | Maintenance of SEXSKINS rows. | CYCLES, REPRO_NOTES, SEXSKINS |
| Social and Multiparty Interactions | |||
| View | One row for each | Purpose | Tables/Views used |
| ACTOR_ACTEES | INTERACT row | Maintenance of social interaction data, INTERACT rows and POINTS. A view optimized for highest performance when working with these tables. Analysis of social interaction data. | INTERACT, PARTS |
| INTERACT | INTERACT_DATA row | Presents INTERACT_DATA with additional date and time columns that transform the underlying date and time columns in useful and interesting ways. | INTERACT_DATA |
| INTERACT_SORTED | INTERACT_DATA row | Presents the INTERACT view sorted in a fashion expected to ease maintenance. | INTERACT_DATA |
| MPI_EVENTS | MPI_DATA row | Analysis and correction of multiparty interaction data. | MPI_DATA, MPI_PARTS, MPIACTS |
| POINTS | POINT_DATA row | Presents POINT_DATA with the Ptime column transformed into a column that may be useful and interesting. | POINT_DATA |
| POINTS_SORTED | POINTS row | Presents POINTS sorted by Sid, and within that by Ptime. | POINTS |
| SAMPLES_GOFF | SAMPLES row | Presents SAMPLES with an additional column Grp_of_focal, which has the group of the focal at the time of sampling. | SAMPLES |
| Darting | |||
| View | One row for each | Purpose | Tables/Views used |
| ANESTH_STATS | unique ANESTHS.Dartid value -- for each darting during which additional anesthetic was administered | Analysis and “eyeballing” of data involving additional administration of anesthetic when darting. | ANESTHS |
| BODYTEMP_STATS | unique BODYTEMPS.Dartid value -- for each darting having body temperature measurements | Analysis and “eyeballing” of darting body temperature measurements. | BODYTEMPS |
| CHEST_STATS | unique CHESTS.Dartid value -- for each darting having chest circumference measurements | Analysis and “eyeballing” of darting chest circumference measurements. | CHESTS |
| CROWNRUMP_STATS | unique CROWNRUMPS.Dartid value -- for each darting having crown-to-rump measurements | Analysis and “eyeballing” of darting crown-to-rump measurements. | CROWNRUMPS |
| DSAMPLES | unique DARTINGS.Dartid value -- for each darting | Visualization of all samples collected per darting. | DARTINGS, MEMBERS, DART_SAMPLES |
| DENT_CODES | unique TEETH.Dartid value -- for each darting with recorded tooth information | Perusal and maintenance of TEETH rows by kind of tooth. | TEETH |
| DENT_SITES | unique TEETH.Dartid value -- for each darting with recorded tooth information | Perusal of TEETH rows by position in the mouth. | TEETH, TOOTHCODES |
| HUMERUS_STATS | unique HUMERUSES.Dartid value -- for each darting having humerus length measurements | Analysis and “eyeballing” of darting humerus length measurements. | HUMERUSES |
| PCV_STATS | unique PCVS.Dartid value -- for each darting having PCV measurements | Analysis and “eyeballing” of darting PCV measurements. | PCVS |
| TESTES_ARC_STATS | unique TESTES_ARC.Dartid value -- for each darting having at least one measurement of testes length or width circumference | Analysis of testes length and width measurements taken during darting. | TESTES_ARC |
| TESTES_DIAM_STATS | unique TESTES_DIAM.Dartid value -- for each darting having at least one measurement of testes length or width diameter | Analysis of testes length and width measurements taken during darting. | TESTES_DIAM |
| ULNA_STATS | unique ULNAS.Dartid value -- for each darting having ulna length measurements | Analysis and “eyeballing” of darting ulna length measurements. | ULNAS |
| VAGINAL_PH_STATS | unique VAGINAL_PHS.Dartid value -- for each darting having vaginal pH measurements | Analysis and “eyeballing” of darting vaginal pH measurements. | VAGINAL_PHS |
| Inventory | |||
| View | One row for each | Purpose | Tables/Views used |
| LOCATIONS_FREE | LOCATIONS row that isn't used in NUCACID_DATA or in TISSUE_DATA | Querying of available ("free") locations for storing new samples | LOCATIONS, NUCACID_DATA, TISSUE_DATA |
| NUCACID_CONCS | NUCACID_CONC_DATA row | Converting and standardizing units of nucleic acid concentration | NUCACID_CONC_DATA, NUCACID_CONC_METHODS, NUCACID_LOCAL_IDS |
| NUCACID_SOURCES_EXT | NUCACID_SOURCES row | Viewing, inserting, and updating data about nucleic acid sources with their LocalId's | NUCACID_SOURCES, NUCACID_LOCAL_IDS |
| NUCACIDS | NUCACID_DATA row | Showing data about nucleic acids in a human-readable format | NUCACID_DATA, TISSUE_DATA, UNIQUE_INDIVS, BIOGRAPH, NUCACID_LOCAL_IDS, NUCACID_SOURCES |
| NUCACIDS_W_CONC | NUCACID_DATA row | Showing data about nucleic acids in a human-readable format, including concentrations from the most-recent quantifications | NUCACID_DATA, NUCACID_CONC_DATA, TISSUE_DATA, UNIQUE_INDIVS, BIOGRAPH, NUCACID_LOCAL_IDS, NUCACID_SOURCES |
| TISSUE_SOURCES_EXT | TISSUE_SOURCES row | Viewing, inserting, and updating data about tissue sources with their LocalId's | TISSUE_SOURCES, TISSUE_LOCAL_IDS |
| TISSUES | TISSUE_DATA row | Showing data about tissue samples in a human-readable format | TISSUE_DATA, UNIQUE_INDIVS, BIOGRAPH, TISSUE_LOCAL_IDS |
| TISSUES_HORMONES | TISSUE_DATA row | Providing an expanded set of information about tissue samples used for hormone analysis. Also useful for simultaneous upload of data to TISSUE_DATA and HORMONE_SAMPLE_DATA | TISSUE_DATA, UNIQUE_INDIVS, BIOGRAPH, TISSUE_LOCAL_IDS, HORMONE_SAMPLE_DATA |
| SWERB Data (Group-level Geolocation Data) | |||
| View | One row for each | Purpose | Tables/Views used |
| QUADS | QUAD_DATA row | Querying of X, Y coodinates from and maintenance of QUAD_DATA rows. | QUAD_DATA |
| SWERB | SWERB_DATA row -- for every SWERB event, departure from camp excluded | Collects SWERB related information spread among several tables and separates geolocation points into X and Y coordinates. | SWERB_DATA, QUADS, SWERB_BES, SWERB_DEPARTS_DATA, SWERB_DEPARTS_GPS |
| SWERB_DATA_XY | SWERB_DATA row -- for every SWERB event, departure from camp excluded | Separates SWERB_DATA geolocation points into X and Y coordinates for ease of maintenance. | SWERB_DATA |
| SWERB_DEPARTS | SWERB_DEPARTS_DATArow -- for every departure from camp of every observation team, for those observation teams which have collected SWERB data | Collects departure related information spread among several tables and separates geolocation points into X and Y coordinates. | SWERB_DEPARTS_DATA, SWERB_DEPARTS_GPS |
| SWERB_GW_LOCS | SWERB_GW_LOC_DATA row -- for every geolocation of an object, of a grove or waterhole | Collects SWERB grove and waterhole location information spread between tables and separates geolocation points into X and Y coordinates. | SWERB_GW_LOC_DATA, QUADS |
| SWERB_GW_LOC_DATA_XY | SWERB_GW_LOC_DATA row -- for every geolocation of an object, of a grove or waterhole | Separates SWERB_GW_LOC_DATA geolocation points into X and Y coordinates for ease of maintenance. | SWERB_GW_LOC_DATA |
| SWERB_LOC_GPS_XY | SWERB_LOC_GPS row -- for every time a group is observed at a geolocated physical object, usually a grove or waterhole, and 2 GPS waypoints are required to by the protocol to collect the data | Separates SWERB_LOC_GPS geolocation points into X and Y coordinates for ease of maintenance. | SWERB_LOC_DATA, ADCODES |
| SWERB_LOCS | SWERB_LOC_DATA row -- for every time a group is observed at a geolocated physical object, usually a grove or waterhole | Presents the relationship between the groups and physical features of the landscape in a more comprehensive manner for simpler querying. | SWERB_LOC_DATA, ADCODES |
| SWERB_UPLOAD | row uploaded into SWERB | This view returns no rows, it is used only to upload data into the swerb portion of Babase. | SWERB_DEPARTS_DATA, SWERB_DEPARTS_GPS, SWERB_BES, SWERB_DATA, SWERB_LOC_DATA |
| Weather Data | |||
| View | One row for each | Purpose | Tables/Views used |
| CURATED_TEMPS | WREADINGS row with a "curated" Tempmin or Tempmax; or for each 06:00 AM - 06:00 AM "day" having any rows in DIGITAL_WEATHER with a "curated" AirTemp_Min or AirTemp_Max | Collation of electronically and manually collected weather data into a single place. | WREADINGS TEMPMINS TEMPMAXS DIGITAL_WEATHER |
| MIN_MAXS | WREADINGS row | Analysis and correlation of manually collected weather data. | WREADINGS TEMPMINS TEMPMAXS RAINGAUGES |
| MIN_MAXS_SORTED | WREADINGS row | The MIN_MAXS view sorted for convienience. | WREADINGS TEMPMINS TEMPMAXS RAINGAUGES |
In addition to the above views there are a number of views
which produce the group of a referenced individual as of a
pertinent date. These views are all named after the table from
which they are derived, with the addition of the suffixed
_GRP. They are nearly identical to the table
from which they derive, differing only by the addition of a
column named Grp. The views which produce an
individual's group are listed in the following table.
Table 2.5. The table_GRP Views
To as great an extent as possible Babase utilizes a controlled vocabulary within the system's data store. Again, as far as is possible, this vocabulary may be tailored by adding or deleting codes to tables that define the vocabulary used elsewhere.[23]
At times, the Babase system recognizes that particular
codes have special meanings, for example, the BIOGRAPH table's
F (female) Sex code or the
0 (alive) Status code. The meaning of these codes is
fixed into the logic of the system. As examples, an individual
must be female to be allowed to have a menstruation, or, the
individual must be alive if a sexual cycle event is to post-date
the individual's Statdate. Some of
these codes, like sex, are not defined in tables, they are
hardcoded into the system. Others are defined in support or
other tables. Because these codes have intrinsic meaning, they
cannot be removed from the Babase system nor should their
presence in the data be used to code a different meaning from
that which the code presently has. For example, the meaning of
STATUSES code value
0 should not be changed to mean
“death due to meteorite impact” because the
system's programs would then allow dead individuals to have
sexual cycles. Each of the “special” values that
the system requires retain particular meaning is listed in the
Special Values section of the table's documentation. For further
information on the meaning of the “special” values,
see the description of the data table(s) that contain the code
values. Should the meaning of one of these
“special” values need to be changed, the logic in
the Babase programs should be adjusted to reflect the
change.
Babase prevents ordinary users from altering rows that
contain special values in an attempt to prevent
mis-configuration of the system. Only users with permissions to
modify a table's triggers may alter the table's special values.
This is not a panacea. To return to the example above, not only
does the system expect a STATUSES code of
0 to mean alive, it also expects
0 to be the
only code on STATUSES
that means alive. If another STATUSES code is
created to indicate a more specific sort of
“alive-ness”, unless re-programmed the system will
consider all individuals given that code to be dead, not alive.
A careful review of the documentation should be undertaken
before modifying the content of tables that instantiate special
values.
Indexes are a feature of databases, a feature which greatly speeds data retrieval. In return there is a small cost in the time it takes to change table content, and cost in disk space used. Databases generally require indexes to perform efficently. It is a good idea to index the tables each user has in their personal schema.
There is no documentation on the indexes used in Babase. In general, there is an index for each way the tables are commonly referenced. For example, if records are often looked up on the basis of date, there will be an index on the date. As a practical guide, there is an index on each of the columns at the endpoint of a “relational line” in the above entity-relationship diagrams, as well as an index on every date column with the exception of the CYCPOINTS table's Edate and Ldate columns. Almost all indexes are b-tree indexes.
Babase uses common and widespread Unix development tools and techniques[24] to minimize a new developer's learning curve. This is a vain hope. Babase is complex and contains a lot of moving parts.
The remainder of this section describes conventions and procedures that those working with the Babase source code are expected to follow. It is of interest primarily to those who work with, or are considering working with, the code. It is not a comprehensive list, guidance should be taken from the existing code.
Anything and everything that is part of Babase should be checked into the project's revision control system.
All data values used in the code should be abstracted,
either via m4 or PHP defines, using names that begin with
“bb_”.
Minimize hardcoding. The use of data values in the code should be minimized. By keeping the number of hardcoded values to a minimum, the values used within the system can be altered through procedural changes alone, expensive programming can be avoided, and the flexibility of the system is increased.[25]
All database extension, triggers, functions, etc. should be written in PL/PgSQL, supplemented by m4.
All stand-alone programs should be accessible via the web. They should be written in PHP and styled with CSS2. The web pages they produce should be XHTML 1.0 compliant and should pass W3C validation at http://validator.w3.org/. Style sheets should pass the CSS validator at http://jigsaw.w3.org/css-validator/. Programs that access the database should obtain their PostgreSQL login credentials from the user, preferably using the existing PHP library code.
Each database user must be assigned unique login credentials to the PostgreSQL database. Each user is responsible for the security of his own login credentials and should never use login credentials that are not her own. All code should support this paradigm.
Every file should begin with a statement of copyright.
Each program, function, or procedure should have documented: its input arguments; its return value; any side effects including changes to pass-by-reference arguments, changes to the screen, changes to the database cursors, etc.
Clarity in your code is more important than efficiency. If the code is not clear, it is less likely to work and more likely to have bugs introduced upon maintenance. There is no point in getting a wrong answer quickly.
See the README files in the source tree's directories for information on how the source code is organized.
[17] As security restrictions permit, of course.
[18] That way if you unknowingly revealed your password to the terrorists last weekend when you were drunk, by the time everybody sobers up the password will have been changed and the amount of damage done is limited.
[19] There is one exception to this rule. Members of the
babase_editors group actually may insert data to these
tables, but only when it is done automatically as part of
an UPDATE or DELETE
in a The babase schema table.
[20] Manual updates probably shouldn't be allowed either, but we need to allow automatic updates resulting from legitimate data changes made by babase editors. To allow this, the rule is that only admins are allowed to update this column at all, and the "versioning" function is always run as an admin.
[21] Don't try this at home! Trained Professionals Only! Etc. ;-)
[22] And the reverse is true. The id of a row in the first table can be used to find the row in the second table that holds it.
[23] Examples may be readily found in the Chapter: “Support Tables”.
[24] Usually.
[25] This is very important but the reasons behind it are not obvious, coding values into the programs means creating office procedures that cannot be altered without a programmer. For example, encoding the value of the unknown group into the system would make it impossible to create different unknown groups for animals disappearing from different groups, or different unknown groups for animals disappearing in varying states of health, or whatever.
Table of Contents
- Group Membership and Life Events
- Physical Traits
- Sexual Cycles
- Social and Multiparty Interactions
- ALLMISCS (Ad-libitum sample data)
- CONSORTS (multiparty disputes over CONSORTshipS)
- FPOINTS (Point data on Females)
- INTERACT_DATA (Interactions)
- MPIS (Multiparty InteractionS)
- MPI_DATA (Multiparty dyadic Interactions)
- MPI_PARTS (Multiparty Interaction PARTicipantS)
- PARTS (Participants in interactions)
- POINT_DATA (Point observation data)
- NEIGHBORS (point observation data on Neighbors)
- SAMPLES (all-occurrences Samples)
- Darting
- ANESTHS (Extra Sedation Administered During Darting)
- BODYTEMPS (Darting Body Temperature Measurements)
- CHESTS (Darting Chest Circumference Measurements)
- CROWNRUMPS (Darting Crown-to-Rump Measurements)
- DART_SAMPLES (Darting Tissue Sample Records)
- DARTINGS (Baboon Darting Events)
- DPHYS (Darting Physiological Measurements)
- HUMERUSES (Darting Humerus Length Measurements)
- PCVS (Darting Blood Measurements)
- TEETH (Darting Tooth Data)
- TESTES_ARC (Darting Testes circumference Data)
- TESTES_DIAM (Darting Testes Diameter Data)
- TICKS (Darting Tick and Parasite Data)
- ULNAS (Darting Ulna Length Measurements)
- VAGINAL_PHS (Darting Vaginal pH Measurements)
- Inventory
- LOCATIONS
- NUCACID_CONC_DATA (NUCleic ACID CONCentration DATA)
- NUCACID_CREATORS (NUCleic ACID CREATORS)
- NUCACID_DATA (General information about NUCleic ACID samples)
- NUCACID_LOCAL_IDS (LOCAL IDentifierS for NUCleic ACID samples)
- NUCACID_SOURCES
- POPULATIONS
- TISSUE_DATA (General information about TISSUE samples)
- TISSUE_LOCAL_IDS (LOCAL IDentifierS for TISSUE samples)
- TISSUE_SOURCES
- UNIQUE_INDIVS (All UNIQUE INDIVidualS)
- SWERB Data (Group-level Geolocation Data)
- AERIALS (Aerial photos)
- GPS_UNITS (Individual GPS Devices)
- QUAD_DATA (map Quadrants)
- SWERB_BES (Begin/Ends: Uninterrupted bouts of group-level observation)
- SWERB_DATA (Group Level GPS Point Samples)
- SWERB_DEPARTS_DATA (Observation team departures from camp)
- SWERB_DEPARTS_GPS (SWERB GPS Departure data)
- SWERB_GWS (SWERB Grove and Waterholes)
- SWERB_GW_LOC_DATA (SWERB Grove/Waterhole Location Data)
- SWERB_LOC_DATA (LOCation-Specific DATA from GPS Points)
- SWERB_LOC_GPS (Secondary Data for LOCations in GPS points)
- SWERB_OBSERVERS
- TREES
- Weather Data
- Metadata
These tables contain the permanent records of baboon-related data. For the most part this data are as collected in the field, although presumably the field staff is not perfect and there will be some errors that are corrected before data entry into Babase. Some columns, and more rarely entire rows, do contain derived data. Some of the derived data, such as pregnancy parity, is manually maintained, other derived data, such as sexual cycle sequence numbers or menses dates computed from onset of turgesence, is maintained by the system. The documentation clearly indicates which data are collected in the field, which data are derivative, and how derived data values are constructed.[26]
This table records cases where short names (Snames) were assigned to individuals and then the choice of name was rescinded. It contains one row for every rescinded Sname, linking the rescinded value to the Sname presently assigned to the individual.
A new row may not be inserted into BIOGRAPH with an Sname value that is an Alternate_Sname value. However, in order to accommodate cases of switched identities, ALTERNATE_SNAME rows may have Alternate_Sname values which appear in the BIOGRAPH.Sname column.
The Sname value must differ from the Alternate_Sname value.
A three-letter code (an id) that uniquely identifies a particular animal (an Sname) in BIOGRAPH. This code can be used to retrieve information from BIOGRAPH or other places where the animal's three-letter code appears.
This column may not be NULL and may not be
998.
An Sname once associated
with the individual identified in the Sname column. This
column may not be empty, it must contain exactly 3
characters, it may not contain lower case letters, and it
may not contain the space character. This column may not be
NULL.
The name associated with the alternate sname. This column may not be empty, it must contain characters, and it must contain at least one non-whitespace character.
This column may be NULL.
Notes regarding the existence of the alternate Sname. This column may not be empty, it must contain characters, and it must contain at least one non-whitespace character.
This column may not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table records the basic biographical data on
baboons. It contains one row for each baboon, including still
births and fetal deaths (collectively, fetal losses), on which
data have been collected. In all cases the Statdate value
must not be less than the Birth value. Live animals, those
with a Status of
0, must have a recorded cause of
death of “not applicable”, a Dcause of
0. Live animals that have no
associated CENSUS rows (absences excepted) must have a
Statdate equal to their Birth date. Animals with no recorded
cause of death, a Dcause of
0, must have “not
applicable” as the degree of confidence in both the
nature and agent of death; their DcauseNatureConfidence and DcauseAgentConfidence must both be
0.
The system will generate an error when it finds a birth date that is later than the the team's last contact with the mother -- when the Birth date is later than the mother's Statdate.[27]
All individuals with an Sname, i.e. those that aren't
fetal losses, must have a Name and will have rows in MEMBERS. Individuals with an Sname
may not have their Sname removed (set to NULL).
Caution
The Psionload program treats an
Sname value of 998 in
a special fashion.
998 may not be used
as an Sname value. See the Psionload
documentation below for details.
Those rows that record data on fetal losses must
maintain the following relations between their data values:
the Sname, Name, Entrydate, and Entrytype values must be NULL; the
Statdate must be the same as the birth date (Birth); and the
Status must not be 0 (alive).
Because fetal losses have no Sname they cannot have
corresponding CENSUS rows and there will not
be any record of their group membership in MEMBERS.
Entrydate and Entrytype can only be NULL for fetal
losses--when their Sname is also
NULL. Otherwise, they cannot be NULL and Entrydate must be between the
individual's Birth and Statdate values, inclusive. When Entrytype is
B (Birth), the Entrydate must be the individual's Birth. When Entrytype is any other value, Entrydate cannot equal Birth.
The Statdate of live individuals is derived from the CENSUS table. An actual census does not have to be taken. Any observation of an individual in a group that results in a row being added to CENSUS is sufficient, except that Absences don't count. When there are no non-absent censuses and the individual is alive, then the Statdate is the Entrydate. This column is automatically updated when CENSUS is updated to ensure that these conditions remain true. When the individual is not alive the Statdate is the date of death.
Caution
Living individuals, unlike dead ones, can have MEMBERS rows created by the interpolation procedure that locate the individual in a group on a date later than the individual's Statdate. For further information see: Interpolation at the Statdate.
In a like fashion, living individuals, unlike dead ones, can have CYCPOINTS rows created by automatic Mdate generation on a date later than the individual's Statdate. For further information see: Automatic Mdate Generation.
Caution
Male Dispersed dates may be after the Statdate when the individual is alive and there are subsequent censuses of the group from which the individual dispersed.
Caution
When dates are encoded as intervals to account for uncertainty in the data, as with the CYCPOINTS Edate and Ldate columns, the latter end of the interval may post-date the Statdate.
Aside from the preceding caveats, Babase does not allow data to be related with an individual when the date of the data postdates the individual's Statdate. Therefore Statdate provides a convenient way of determining the end of the time interval during which there are data on an individual, a way that is independent of whether the individual is alive or dead.
An individual's Dcause represents a specific Nature and Agent of death. When considering the associated DcauseNatureConfidence and DcauseAgentConfidence values, it is important to remember that a Dcause should be interpreted as "if Nature, then Agent". It is tempting to assume that this means that the DcauseAgentConfidence cannot be higher than the DcauseNatureConfidence, but this is not so. The DcauseAgentConfidence is assigned contingent on the associated Nature being true, so it is possible for the DcauseAgentConfidence to be higher than the DcauseNatureConfidence. For this reason, the system has no rules validating the DcauseAgentConfidence based on the DcauseNatureConfidence, nor vice versa.
Confidence in the accuracy of the estimated birth date is categorized in the Bstatus column. The estimated range of possible birth dates might not be as symmetrical around the Birth date as is implied in BSTATUSES, so the specific boundaries of this range are recorded in the EarliestBirth and LatestBirth columns.
The EarliestBirth and LatestBirth columns cannot be NULL,
unless the Bstatus is
9.0 ("unknown"), in which case
both EarliestBirth and LatestBirth must be NULL.
The EarliestBirth must be on or before the individual's Birth, which must be on or before the individual's LatestBirth. LatestBirth must be on or before the individual's Statdate, but only for individuals with non-absent rows in CENSUS.[28]
The LatestBirth must be on or
before the Entrydate, unless the
individual's Entrytype is
B (Birth). As mentioned
above, when Entrytype is
B, the Entrydate must equal the Birth date. In these cases, if there is
any uncertainty about when the
individual's "true" birth date is, the LatestBirth might legitimately be after
the Birth date and therefore after
the Entrydate. The LatestBirth should never be long after
the Entrydate[29], so even in these cases there are boundaries
placed on LatestBirth. When Entrytype is
B (Birth), the LatestBirth cannot be more than
29
days[30] after the Entrydate
unless either or both of them is NULL.
The system will return a warning if the length of time
between EarliestBirth and LatestBirth is more than Bstatus years[31]. Similarly, the system will return a warning if
the EarliestBirth is more than
(0.5 × Bstatus) years before the Birth date, and another if the LatestBirth is more than
(0.5 × Bstatus) years after the Birth date.
It's possible for an individual's Bstatus to be "too high", based on the length of time between EarliestBirth and LatestBirth. That is, a high Bstatus could mistakenly be used for individuals whose EarliestBirth - LatestBirth range is significantly less than Bstatus years. The system will return a warning if the length of time between the EarliestBirth and LatestBirth is less than or equal to the length of time indicated by a smaller BSTATUSES.Bstatus value.
When inserting or
updating data in this table, the system can use the row's
Bstatus to automatically populate
the EarliestBirth and LatestBirth columns, if desired. When the
Bstatus is not
9.0 ("unknown"):
Any provided EarliestBirth or LatestBirth values are inserted into their corresponding columns.
When no EarliestBirth is provided, this column is calculated as the Birth date − (
0.5× Bstatus years).When no LatestBirth is provided, this column is calculated as the Birth date + (
0.5× Bstatus years).
While the UNIQUE_INDIVS table contains a larger list of
all individuals across multiple
populations, this table is the primary authority for the
"main" population. When rows are inserted or deleted in this
table, related rows are automatically inserted or deleted in
UNIQUE_INDIVS with IndivId = the Bioid, and PopId =
1. Individuals in
the main population cannot be added to UNIQUE_INDIVS before being added to this
table.
A unique integer identifying the BIOGRAPH row.
Babase rarely uses this identifier; it exists for the convenience of application programs and for distinguishing individuals without Snames (fetal losses) from each other and from other individuals.[32]
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The short name of the individual. This is an exactly three character long name abbreviation which is used to identify the individual and so must be a unique data value. It may not contain lower case letters or spaces.
Tip
The Sname is usually, but not always, the first 3 characters of the Name.
This value appears in many other places in the system and
so should not be changed without changing all the other
places in the database where the abbreviation appears;
really, once established, the only reason to change this
column is because the short name had already been
used. [33] Because this is unlikely, Babase does not
allow the Sname to be changed. The Sname is always
composed of capital letters and may not contain a
space.[34] This column should only
be NULL if the row represents a fetal loss.
The name of the individual. This is a textual column
used for descriptive purposes. This value must be unique
when a comparison is done in a case insensitive
fashion. This column should only be NULL if the row
records a fetal loss. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The Pid value, from the PREGS
table, of the individual's mother's pregnancy that ended
in the birth[35]of the individual. This column may be
NULL. A NULL value indicates there is no record of
the individual's mother.
Caution
More than one individual may have the same Pid, as long as they were products of the same pregnancy. This occurs when twins are born into the study population.
This column may not be empty, it must contain characters, and it must contain at least one non-whitespace character.
The sex of the individual. The legal values are:
| Code | Description |
|---|---|
| M | the individual is male |
| F | the individual is female |
| U | the individual is of unknown sex |
This column may not be NULL.
The date the pregnancy ends. For live births and for individuals whose maternity is unknown, this is their estimated birth date. Otherwise, this is the date of the fetal loss. (A pregnancy that ends with the mother's death is considered as a spontaneous abortion (fetal loss) for this purpose.)
This column may not be NULL.
The BSTATUSES.Bstatus categorizing the quality of the birth date estimate.
This column may not be NULL.
The maternal group of the individual, the Gid of the group into which the individual was born.
This column must contain a Gid value of a row on the
GROUPS table. This column
may not be NULL.
Tip
If the maternal group is not known, the maternal group should be recorded as the unknown group.
Matgrpconfidence (confidence in maternal group assignment)
The degree of confidence in the assignment of the Matgrp value. The legal values for this column are defined by the CONFIDENCES support table.
This column may not be NULL.
The date the individual entered the study population.
Note
Because of Interpolation, it may seem like this column could be maintained automatically. However, the opacity of "non-interpolating" rows in CENSUS and the related historical analyses prevent accurate automatic determination of the entry date for many individuals. For more information, see CENSUS.Status and Interpolation, Data are not Re-Analyzed.
This column can be NULL, only if the row
represents a fetal loss.
The way the individual entered the study population. The legal values for this column are defined by the ENTRYTYPES table.
This column can be NULL, only if the row
represents a fetal loss.
The status date of the individual. When the individual is alive, this is the latest date on which the animal was censused and found in a group.
This column may not be NULL.
The state of the individual's life at the Statdate. The legal values for this column are defined by the STATUSES support table.
This column may not be NULL.
The cause of death or circumstances associated with death. The legal values for this column are defined by the DCAUSES support table.
This column may not be NULL.
The degree of confidence in the nature of the individual's death or circumstances associated with the individual's death (their DCAUSES.Nature). The legal values for this column are defined by the CONFIDENCES support table.
This column may not be NULL.
The degree of confidence in the agent of the individual's death or circumstances associated with the individual's death (their DCAUSES.Agent). The legal values for this column are defined by the CONFIDENCES support table.
This column may not be NULL.
A boolean value indicating whether or not there exist rows on the ALTERNATE_SNAMES table related to the individual's Sname. This value is true if and only if there exists a row on ALTERNATE_SNAMES with an Sname value which is the individual's sname or there exists an ALTERNATE_SNAMES row with a Alternate_Sname value which is the individual's sname.
The value in this column is automatically maintained
and will never be NULL.
The earliest estimated birth date for this individual.
The values in this column may be calculated automatically, as discussed above.
This column may be NULL, but only when the
accuracy of the birth estimate is unknown (when Bstatus is
9.0).
The latest estimated birth date for this individual.
The values in this column may be calculated automatically, as discussed above.
This column may be NULL, but only when the
accuracy of the birth estimate is unknown (when Bstatus is
9.0).
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
The population census table. Aside from the BIOGRAPH.Matgrp column, this table is the origin of all information regarding group membership. This table holds all the field census data and any information regarding group membership that is recorded in the field demography notes. It contains one row per animal per group per day censused. There is an additional row per individual per demography note for those days when there is a demography note regarding the individual and group but no census of the group. (See DEMOG.)
Tip
One way to have Babase record that an individual is alone is to first create a row in GROUPS meaning alone, and then to assign individuals who are alone to this group. The “alone-ness” of an individual can then be tracked in the same fashion as group membership, although the Babase user does then need to be aware that the members of the “alone” group are not actually proximate to one another.
The system will report individuals who are first censused in a group other than their maternal group (BIOGRAPH.Matgrp). The exceptions to this are when the maternal group is the unknown group or that first census row records an absence.
The system will report individuals with a BIOGRAPH.Sname that do not have any related (non-absent) CENSUS rows.
The Date must be between the
Grp's related GROUPS.Start and Cease_To_Exist, inclusive, with one
exception. Rows indicating absences — rows whose Status is A
— may occur outside of the date range for a group's
lifetime. These may sometimes be needed during fission/fusion
periods to manually prevent an individual from being
interpolated into a group that no longer exists or doesn't yet
exist. However, a need for such absences is rare, so the
system will report a warning for any "absent" censuses before
the Grp's Start or after its Cease_To_Exist, exclusive.
The system will report a warning when CENSUS rows have a Status
of C or
D and a Date before the individual's LatestBirth, and another warning if
before the individual's Entrydate.
As noted in the MEMBERS documentation, Babase does not allow an individual to be in more than one group on a given day.
Ideally, the original field census data sheets could be recovered from CENSUS, but there are several situations where that is not possible:
First, a datum is lost when an individual is actually censused in two groups on the same day because of movement between groups and the timing of the censuses.[36] In this situation a decision should be made as to which group CENSUS should record the individual's presence on that day. A demography note should then be added to DEMOG, with text that notes the individual's presence in the second group. This results, technically, in all of the information from both censuses, or other location information, being entered into the database. However, it should be remembered that, because the information regarding the second census is in textual form, it is not readily available to automated tools.
Second, it may be necessary during group fissions and fusions to record a different Grp than what was actually recorded because it is usally not clear in real time that a fission/fusion has begun. There is necessarily a lag between when a change can be seen retroactively and when the field notebooks are actually updated to reflect the existence of the newly-formed group(s). For fusions it is important to construct group membership in Babase carefully, for the sake of maintaining group residency. If an individual is a resident of one parent group and is censused in another, the residency algorithm recognizes the other parent group as an entirely different group. That is, it does not recognize that the groups will soon be related. To prevent a loss of residency due to an apparent group change, censuses in the other parent group(s) should be recorded with the daughter group as the Grp whenever at least some of both parent groups are together.
Example 3.1. Crossovers during a fusion
"Bruce" has been a resident of the "Gotham" group for years. "Clark", meanwhile, is a resident of the "Metropolis" group, and "Diana" is the alpha female (so, definitely resident) of the "Amazons" group. On 01 June, the three will permanently fuse together and form the "JLA" group, after first being seen together on 01 May. (JLA's Start is 01 May, Permanent is 01 June) Throughout May, census records show Bruce making short visits to Metropolis and to the Amazons. Knowing that the groups are in a fusion period, whenever Bruce is with Metropolis or the Amazons, he and all members of the group he is with should be recorded as being in the JLA. Similarly on dates later in the month when Bruce and his close associates Robin and Alfred — along with Clark and sometimes his sister Kara — were with the Amazons, all members of the Amazons and their friends from Gotham and Metropolis should be recorded in the JLA.
In January of the same year, Clark made a brief visit to Gotham. That was before the fusion began in May, so that visit's Grp need not be changed in any way[37].
Third, some CENSUS rows are derived from analyses of historical data and employ MEMBERS-style rows where group members generally have a row on every date of a given month that they were present, rather than just those dates when censuses were performed. See the Status column for details.
Caution
Be careful when changing these data. When CENSUS data are inserted, deleted, or updated, the MEMBERS table and BIOGRAPH.Statdate column are automatically updated via Interpolation. Also, remember that rank will almost certainly change should group membership change.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row. Cenid links CENSUS to DEMOG.
This column may not be NULL.
The date of the census, or the date of the
demography note (when Status is
D).
Note
The date value must not be more than a year later than the present moment. This rule prevents accidental data entry errors from creating so many rows in MEMBERS that all available disk space is used.
This column may not be NULL.
The individual whose location is being recorded. The three-letter code that uniquely identifies an individual in BIOGRAPH. There will always be a row in BIOGRAPH for the individual identified here.
This column may not be NULL.
The group where the individual is located. This is a Gid value from GROUPS. This column should contain the most specific sub-grouping available -- subject to the constraints of the data entry protocol, of course. Aggregation into larger groupings is accomplished by retrieving the associated MEMBERS.Supergroup of the individual on the date of census.
This column may not be NULL.
Note
Usage exception: For dates between 21 Mar 1990 and 29 Feb 1992, inclusive, the group recorded for the sub-groups of Alto's group do not necessarily reflect the actual groupings of the animals on a particular day, but are instead indications of the group-splitting process. See the Protocol for Data Management: Amboseli Baboon Project document for further explanation.
A one letter code indicating the source of the
location information. Status is the source of MEMBERS.Origin data. The current codes are as
follows: C (census),
A (absent),
D (demography), and
M or
N (manual). Other
values derived from analysis of historical data include:
S, E,
F, B,
G, T,
L, and
R.
The CENSUS.Status Codes
C(census) The animal was found in the group on a field census sheet: from the census datasheets. (There may or may not be a corresponding demography note on DEMOG as well.)
Tip
A
CStatus is marked on the field census data sheet as an “X”.A(absent) The animal was not found in the group on a field census sheet. Note that while an individual should not be recorded “present” in more than one group on the same day, s/he may be absent from several groups on any given day.
Tip
An
AStatus is marked on the field census data sheet as an “0”.D(demography) The animal was noted, in the field notebooks or elsewhere, to be in a group but was not marked present in a field census of a study group on that day.[38] There should be a DEMOG row associated with the CENSUS row. The individual may or may not have been marked “absent” on the same group's field census for the day.[39]
Tip
A
DStatus is marked on the field census data sheet as an “0”, when there exists a corresponding place on the census data sheet.Warning
The system will allow CENSUS rows with a Status of
Dto be entered without there being a corresponding DEMOG row in existence.[40] However it is expected that these rows exist only long enough to allow entry of a related DEMOG row. The system will report CENSUS rows with a Status ofDthat have no related DEMOG row.M(manual, interpolated) This code provides a way to manually supplement what is in the CENSUS table when there is no other way to get the data in. Babase considers this code to be the same as the
Ccode.N(manual, not interpolated) This code provides an alternative way to manually supplement what is in the CENSUS table when there is no other way to get the data in. This code does not interpolate, it is presumed to be the result of some analysis.
S(Susan's data) The data comes from the old DISPERSE database where the record had both a Datein and a Dateout.
E(ending date) The data comes from the old DISPERSE database where the record had a Datein but not a Dateout.
F(final date) The data comes from the old DISPERSE database where there is a Dateout and the last recorded location is before the Statdate.
B(birth date) The data comes from the old DISPERSE database where the record had a Dateout but not a Datein.
T(total) The data comes from the old DISPERSE database where the record had neither a Datein nor a Dateout.
G(gap) The data are a record of the animal in the unknown group when the animal appeared in the old DISPERSE database but where there was a gap between times of recorded location.
L(lineage) The group is from the Matgrp on the old CYCTOT database, either because the animal did not appear in the DISPERSE database, or because the first location for the animal in the old DISPERSE database had a Datein and this Datein was after the birth date of the animal.
R(result of Alto's breakup) The datum is
S,E,F,B,G,T, orLdatum that has had locations which were changed from 1.0 to the group in which the animal was censused on 15/4/92. This change left allRrows as part of a contiguous series of days during which the animals are located in the Alto's sub-group as censused on 15/4/92, and the time-adjacent locations were not 1.0.
This column may not be NULL.
Cen is whether or not the CENSUS row represents an entry on
a field census data sheet. TRUE means the CENSUS row exists because of an
entry on a census data sheet, FALSE means there was no
census done and the CENSUS
row exists to support a demography note, manual notation
of absence, etc. Cen should only be TRUE when Status is
C,
A, or
D.
This column may not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table records the dates of first consortship for males; this is a maturational milestone in males that we have analyzed in several contexts. It contains one and only one row for every individual for which there is a recorded first consortship. Individuals who have not yet consorted, or individuals that have consorted but whose first consortship date is not known, do not appear in the table.
Tip
Currently it only contains values for males; females may be added if desired.
Tip
All dates are exact, no “BY” dates are entered as we do for MATUREDATES and RANKDATES, so there is no “Status” column.
When there is a row in this table there must be a sexual maturity date in MATUREDATES, and the consortship date must be later than the sexual maturity date. The Consorted date cannot be before the individual's Entrydate, nor after the individual's Statdate. The individual must be at least 5 years of age on his Consorted date. The system will report a warning if the individual is 12 or more years of age on his Consorted date.
A three-letter code (an id) that uniquely identifies
a particular animal (an Sname) in BIOGRAPH. This code can be used
to retrieve information from BIOGRAPH or other places where
the animal's three-letter code appears. This column may
not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table holds the text that records group membership information not written on the regular field census sheets, especially that from the field demography notes. DEMOG provides a means of notating CENSUS rows, and thus facilitates management of additional “free form” CENSUS rows, rows that do not directly correspond with the field census sheets.[41] Thus, in conjunction with these corresponding CENSUS rows, the DEMOG rows capture group membership information that otherwise would not appear in the CENSUS table.
DEMOG contains one and only one row for every individual for every date for every group where the individual was noted present in free form textual field notes or other miscellaneous sources. The DEMOG row holds textual information. There is always exactly one corresponding CENSUS row, which holds the corresponding group membership information in the usual coded and structured form. (Note that only some CENSUS rows will have DEMOG rows; CENSUS rows that originate entirely in the regular censuses of groups will not, in general, have an associated DEMOG row). A single field note referring to more than one individual must appear in DEMOG as two (or more) separate rows, one row per individual. Multiple field notes pertaining to a single individual on a single date must be combined into one piece of text and entered in a single DEMOG row. (See the Protocol for Data Management: Amboseli Baboon Project for structure of the demography data as entered by the operator.)
Adding or removing DEMOG rows automatically updates the CENSUS.Status column of the corresponding CENSUS row.
Tip
Use the DEMOG_CENSUS view to upload datasets into this table. Use CENSUS_DEMOG view to maintain this table by hand.
Caution
The data integrity rules require that when a demography note is entered the CENSUS row be created before the related DEMOG row.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row. Cenid links CENSUS to DEMOG.
This column may not be NULL.
A code that identifies the written field notebook or other source where the demography note can be found.
The legal values for this column are defined by the
DEMOG_REFERENCES support table, see
below. This column may not be NULL.
The demography note text pertaining to the CENSUS row with the given Cenid.
This column may be NULL.[42]This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table records dates of dispersal for males (females do not disperse and do not appear in this table). It contains one and only one row for every male who has a known date of dispersed from the study groups. Males who have not yet dispersed do not have a row in this table. Only males can have rows on this table.
Tip
All dates are exact, no “BY” dates are entered as we do for MATUREDATES and RANKDATES, so there is no “Status” column.
The system will report a warning when there is a row in
this table and there is no sexual maturity date in MATUREDATES. The Dispersed date must be on or
after the individual's Entrydate.
The Dispersed date cannot be after the individual's Statdate when the individual is not alive
(when BIOGRAPH.Status is not
0). When the individual is alive
the Dispersed date may only be after the Statdate when the individual has been
censused absent (CENSUS.Status is A)
in the group[43]
and the Dispersed date is not after the earliest such
post-Statdate census date.
The system will returning a warning when the Dispersed date is before his LatestBirth.
A three-letter code (an id) that uniquely identifies a
particular animal (an Sname) in BIOGRAPH. This code can be used to
retrieve information from BIOGRAPH or other places where the
animal's three-letter code appears. This column may not be
NULL.
The degree of confidence in the assignment of dispersal date or rationale behind the assignment of the dispersal date. The legal values for this column are defined by the CONFIDENCES support table.
This column may not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every group on which
there is some recorded information. This includes not only the
study groups and non-study groups, but also temporary
daughter groups and the special group “Unknown”[44](See the Protocol for Data
Management: Amboseli Baboon Project for when to use this special
group.) When a daughter group becomes a regular group (after a
fission or fusion is complete), the new group should be given
a Permanent date to indicate that it
is now a permanent group (Permanent
is not NULL). Any “old” daughter groups that
did not become permanent should be left in GROUPS to support
the daughter grouping membership history.
Tip
This table serves primarily as a tool for the system for data validation. To see its contents in a more human-readable format, use the GROUPS_HISTORY view.
Every reference to a group elsewhere in the Babase
system corresponds to a Gid
of one of the records in this table. Temporary groups (those
with Permanent of
NULL) must have a
non-NULL> From_group value. Permanent
groups must not have a Permanent
value that is earlier than their Start value. Permanent
groups may or may not have a NULL From_group value.
Note that there is no particular reason to remove from GROUPS those daughter groups that exist for only a short time during group fission. Those sorts of groups can remain temporary forever.
Tip
The MEMBERS.Supergroup column may be used to determine the supergroup of an individual on any given date.
Neither a GROUPS row's From_group value nor its To_group value may be the same as its Gid value.
A group's Permanent and From_group cannot both be NULL. But both
can be non-NULL.
The Cease_To_Exist value must
be NULL or greater than the Start value. The Study_Grp value must be NULL or must not
be less than the Start value. When the
Cease_To_Exist
and the Study_Grp value are both
non-NULL the Study_Grp value must
not be after the Cease_To_Exist
value.
The Cease_To_Exist value must also be greater than or equal to all daughter groups' Start values.
The Last_Reg_Census value must
be NULL or greater than the Start
value. It also must be less than or equal to the group's
Cease_To_Exist date, unless the
Cease_To_Exist is also NULL. And Last_Reg_Census must be NULL or Study_Grp must be NULL or Last_Reg_Census must be on or after the
Study_Grp date. The Last_Reg_Census must be NULL when Study_Grp is NULL.
The Cease_To_Exist must be the
day preceeding the Permanent date of
any daughter groups, unless the daughter group's Permanent is NULL. An important
consequence is that all of a group's permanent daughter groups
must have the same Permanent
date.
A group that is a fusion product cannot have a fission
parent -- the From_group must be
NULL when the group is the result of group fusion, i.e.,
when the group's Gid appears in the
To_group column of another group.
[45]
Caution
The system enforces the rules of the 3 previous
paragraphs "on-commit". In a transaction ending with a
ROLLBACK, any changes to this table will
not be validated against these rules. This means it is
possible for an invalid change to appear error-free if
executed in a rolled-back transaction. Committed
transactions (and commands executed outside of transactions)
perform this check as expected.
The One_letter_code value must be unique within the time period from the group's Start date through the group's Cease_To_Exist date, inclusive of endpoints.
Individuals cannot be placed into rows in the CENSUS table before the Start date of the group, or cannot be
censused in the group at all if the value of the Start column
is NULL. Individuals cannot be placed into rows of the
CENSUS table after the Cease_To_Exist value of the group. Note
that both these restrictions apply to all CENSUS rows, even those that indicate the
individual is absent from the group.
Gaps in observation of a group cannot be added to the BEHAVE_GAPS table if the Gap_Start or Gap_End are before the Start date of the group. Similarly, gaps cannot be added to BEHAVE_GAPS if the Gap_Start or Gap_End are after the Cease_To_Exist date.
Warning
Some gaps in BEHAVE_GAPS may have a Gap_Start date that is equal to the group's Start or Permanent date, implying that the gap started because of the opening of observation of the group.[46] Gaps may also have a BEHAVE_GAPS.Gap_End date equal to the group's Last_Reg_Census or Cease_To_Exist date, implying that the gap ended because of the group's end.[47] If the Start, Permanent, Last_Reg_Census, or Cease_To_Exist column is updated, then these implications will no longer be true. The system makes no attempt to judge whether these implications really are true or just coincidence, so data managers must exercise this judgment. When changing any of these dates in GROUPS, be sure to check for rows in BEHAVE_GAPS with Gap_Start or Gap_End dates that also should be updated, and correct them as needed.
Group
9.0,
Unknown, has a special meaning.
Individuals are placed in this group by Interpolation when their
whereabouts are unknown. Also, a SWERB_DATA.Seen_grp
value of 9.0 in rows with an
Event value of
O indicates an exceptional
circumstance where Seen_grp is
allowed to equal the related SWERB_BES.Focal_grp
value. Another group code for unknown whereabouts should
not be created.
The 10.0 group has the
special meaning of “lone animal”. The SWERB_UPLOAD view uses this value as the SWERB_DATA.Seen_grp
when a lone animal is sighted. Another group code for lone
animals should not be created.
The 99.0 group has the
special meaning of “predator sighting”. The
SWERB_UPLOAD view uses this value as the
SWERB_DATA.Seen_grp when a predator is sighted.
Another group code for predator sightings should not be
created.
A positive numeric value with six digits (4 decimal
places) that identifies the group. Each Gid must be
unique. This column may not be NULL.
The spelled out name of the group. This column must
be unique, and unique insensitive of case. This column may
be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The Gid of the group from
which this group split off, if the group is a fission
product. This column may be NULL.
The Gid of the group formed when the daughter of the group is the result of group fusion.
This column may be NULL to indicate there is no
daughter group or the daughter groups are fission
products.
This column contains the date the group became a
permanent, regular group, or contains NULL if it has not
and is a temporary daughter group. For groups that were
created as a result of fissions or fusions this
column represents the end date of the fission/fusion
period. For groups that were already intact when
observation began this column represents the
first day of observation on that group.
Note
Permanent affects whether or not an individual can be censused only in a daughter group and still be ranked in the parent supergroup. See RANKS and MEMBERS.Supergroup for further information.
The date the group came into existence (or the
earliest date it must have existed in the case of those
groups existent before they were monitored.) The value of
this column may be NULL to indicate the group exists but
is not monitored.
If any parent group has the daughter group as its To_group then the start date is also the date the fusion started.[48]
The date on which the group is deemed to have
permanently dissolved into fission products or merged into
a fusion product. This column may be NULL for groups
still under observation, groups that have not yet
dissolved/merged, and groups whose dissolution/merge
occurred while not under regular observation.
The date of the last regular census done on the
group for study groups that were dropped or ceased to
exist because of fission/fusion. This column may be NULL
if the group hasn't been dropped or was never a study
group.
A 3 character, and exactly 3 character, code that
uniquely identifies the group. The characters must all be
upper case. This code is used by the Psion data
collection devices and in SWERB observations taken using
handheld GPS units and exists solely as a cross reference
from those devices to the regular Babase group Gids. This
column may be NULL if the group is never monitored using
the Psion devices or SWERB GPS devices.
A 1 character, and exactly 1 character, code that
uniquely identifies the group within the time period of
the groups existence. The character must all be upper
case. This code is used to cross reference SWERB waypoint
data to the regular Babase group Gids. This column may be
NULL.
The date the group first became an "official" study
group[49] or NULL if the group was never a study
group.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table records sexual maturity dates, the dates of menarche or testicular enlargement. It contains one and only one row for every animal who matured in a study group or who lived in a study group as a sexually mature individual, and it may occasionally contain a row for a male who was known to mature but who did not live in a study group. Individuals who have not yet matured do not have a row in this table. All sexually mature individuals should have a row in this table. Entry into sexual maturity is not always an obvious or definite event[50], especially for males, so the Matured may be recorded as the first of the month in which the individual entered maturity.
There are restrictions on when an individual may become
mature. The age of an individual at sexual maturity (Matured) must be at least
1016 days. This is about 2.7 years of age.
The system will issue a warning when the sexual maturity
occurs on or before the 3rd birthday. Individuals with a
Mstatus of
O (On) must be mature before
2922 days of age (8 years). The system will
issue a warning when the sexual maturity occurs on or after
the 7th birthday. An individual's sexual maturity date must
be on or before his Statdate.
Some maturity dates are based on irregular observations of individuals before the long-term study began, or before the individuals entered an "official" study group. Either way, these individuals' Matured dates may be long before their Entrydate. Because of this, the system will allow but issue a warning when the month of the maturity date is earlier than the month of the individual's entry into the study population (their Entrydate).
For females, when Mstatus
is O (On) Matured must be the first
T date recorded in the
female's sexual cycling data in the CYCPOINTS table. When Mstatus is not
O Matured may not be after the first
Tdate.
Caution
Changing a female's first Tdate can automatically change the female's Matured date. See CYCPOINTS.
A three-letter code (an id) that uniquely identifies
a particular animal (an Sname) in BIOGRAPH. This code can be used
to retrieve information from BIOGRAPH or other places where
the animal's three-letter code appears. This column may
not be NULL.
This is the date of menarche for females and the
date of testicular enlargement for males, when either of
these dates are known. Otherwise, this is the date by
which the individual is considered to be sexually mature.
See the Protocol for Data
Management: Amboseli Baboon Project for more information regarding the
dates used when the transition to maturity was not
observed.[51] This column may not be NULL.
The status of the maturity date, that is, its
precision, accuracy, quality, or other pertinent
characteristics when it comes to the use of the value.
The legal values for this column are defined by the MSTATUSES support table, see
below. This column may not be NULL.
Tip
This column records whether the animal became mature ON a given (known) date, or BY a given (known) date. If a date is designated as an “ON” date[52] then we are saying that we know the animal attained that marker ON that date.[53] If a date is designated as a "BY" date the animal was adult or subadult BY that date but we do not know when the individual attained it. This scheme allows easy identification of which animals are infants or juveniles on any given day and which are not.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table records dates individuals first attained adult rank. It allows one and only one row for every individual who has attained adult rank. Individuals who have not yet obtained adult rank do not have a row in this table.
The system will report a warning when an individual has a rank (in RANKS) before their Ranked date that is higher (where 1 is highest) than another individual who has already attained adult rank.
Tip
RANKDATES currently contains only data for males but data for females may be added.
When there is a row in this table there must be a sexual
maturity date in MATUREDATES. When MATUREDATES.Mstatus
is O (On) then the rank attainment
date must be later than the sexual maturity date. Otherwise,
the rank attainment date must not be before the sexual
maturity date. The Ranked date cannot be after the
individual's Statdate. All
individuals must be 5 or more years of age
on their rank attainment date. Individuals with a Rstatus of
O (On) must be less than
12 years of age on their rank attainment
date. The system will report a warning for any males over
8.5 (exclusive) that have not yet attained
adult rank.
It is possible that an individual will be known to have attained rank in a non-study group before they entered the study population (their Entrydate). Because of this, the system will allow but issue a warning if an individual's Ranked is before the first of the month of his Entrydate.
A three-letter code (an id) that uniquely identifies
a particular animal (an Sname) in BIOGRAPH. This code can be used
to retrieve information from BIOGRAPH or other places where
the animal's three-letter code appears. This column may
not be NULL.
The date the individual first attained a rank among
adults. The date must fall on the first of the
month. This column may not be NULL.
The status of the rank date, that is, its precision,
accuracy, quality, or other pertinent characteristics when
it comes to the use of the value. The legal values for
this column are defined by the MSTATUSES support table. This
column may not be NULL.
Tip
The legal values for this column are O (for ON) and B (for BY), as with Mstatus in the MATUREDATES table above.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This section contains data about specific physical traits of the individuals.
In general, the data in this section tends to be collected "globally". That is, the data are collected for all living individuals in the study population (or as close to "all" as is possible). While many of the Darting tables could also easily be considered "physical traits", those data are only collected during dartings and therefore are not "global" but instead only available for individuals in that fraction of the population that has been darted.
Records which body parts were affected in each related wound or pathology Cluster, and the quantity of these wounds/pathologies affecting the specific part when that quantity is known. This table contains one row for each recorded body part, per associated wound/pathology (from WP_DETAILS) cluster. For example, if a report indicates two clusters affecting body part A and another cluster affecting body part B, this will be recorded in three rows in this table: two for body part A and one for body part B.
Each WPDId-Bodypart pair must be unique; a wound/pathology cluster can be associated with a particular body part only once.
The Quantity_Affecting_Part column
records the quantity of individual wounds/pathologies in the
related cluster that are affecting this row's body part. When
this quantity is unknown or unclear from the report, or when
the related wound/pathology is not obviously countable
(e.g. "fatigue"), this column should be NULL. When a single
wound/pathology affects more than one body part, this
wound/pathology will be counted more than once: the Quantity_Affecting_Part column
should be 1 for each of the affected parts'
separate rows. For example, if there was a long
slash/laceration extending from the arm to the trunk, this
would be recorded with a Quantity_Affecting_Part of
1 in both the "arm" row and the "trunk"
row, effectively counting a single wound twice.
Warning
Remember, the Quantity_Affecting_Part column indicates the number of wounds/pathologies that were affecting the specified body part. When aggregating data across multiple rows (e.g. sum, average, etc.), remember that individual wounds/pathologies affecting multiple body parts will be counted in more than one row of this table. Using this column to count the number[54]of discrete, independent wounds/pathologies may overestimate the true number.
Tip
When adding or updating data in this table, use the WP_DETAILS_AFFECTEDPARTS view. It is includes related columns from BODYPARTS to facilitate easy entry of Bodypart values, and from WP_DETAILS to determine the appropriate WPDId.
Tip
Use the WP_HEALS or WOUNDSPATHOLOGIES views to select data from this table. These views include related data from the other wound/pathology tables, respectively with and without related healing updates.
A unique identifier for the row. This is an automatically generated sequential number that uniquely identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The WP_DETAILS.WPDId of the wound/pathology associated with this body part.
This column may not be NULL.
A positive integer indicating how many wounds/pathologies of the related type are affecting this body part.
This column may be NULL, when the quantity is
unknown, unclear, or uncountable.
When not NULL, this column cannot exceed
9[55].
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for each cluster of wounds or pathologies that are indicated in a report.
Similar to our use of "report" in WP_REPORTS, a "cluster" of wounds/pathologies is mostly just a data management term: useful for bookkeeping but lacking biological relevance. For our uses, a "cluster" is a group of one or more co-occuring wounds/pathologies of the same type (same WoundPathCode). They are "co-occurring" in that these wounds/pathologies were observed to appear on the same date and were likely not acquired independently. The decision to divide multiple wounds/pathologies into separate one-wound/pathology clusters or to group them into clusters of multiple wounds/pathologies is mostly made by the data manager.
In many cases, how exactly to cluster a set of wounds/pathologies is not a decision, but a necessity. When multiple wounds/pathologies of the same type are indicated on a report, there may be particular MaxDimension, ImpairsLocomotion, and/or InfectionSigns values that apply to some but not all of the wounds/pathologies (e.g. "This one slash is impairing locomotion, those other three are not"). In these cases, it is necessary to divide the multiple wounds/pathologies into separate clusters.
Clusters are numbered in the Cluster column, which must be unique per WPRId.
Some WoundPathCode values
may inherently imply that the ImpairsLocomotion or InfectionSigns column(s) be a particular
value. For example, if an individual is limping, by definition
this pathology is impairing the individual's locomotion so the
ImpairsLocomotion value should
always be Y for that pathology. Because of this
possibility, some validation of the ImpairsLocomotion and InfectionSigns columns is controlled by
values in the WP_WOUNDPATHCODES table and
its ImpairsLocomotion and
InfectionSigns columns.
See the WP_WOUNDPATHCODES documentation for
more details.
The system will return a warning for any WP_DETAILS rows that do not have at least one related row in WP_AFFECTEDPARTS.
Tip
When adding or updating data in this table, use the WP_DETAILS_AFFECTEDPARTS view. It facilitates inserting or updating data with the related WId instead of the WPRId.
Tip
Use the WP_HEALS or WOUNDSPATHOLOGIES views to select data from this table. These views include related data from the other wound/pathology tables, respectively with and without related healing updates.
A unique identifier for this row. This is an automatically generated sequential number that uniquely identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The WP_REPORTS.WPRId of the report in which this wound/pathology cluster was recorded.
This column may not be NULL.
The WP_WOUNDPATHCODES.WoundPathCode for this wound/pathology cluster.
This column may not be NULL.
A positive integer identifying this cluster of wounds/pathologies.
This column may not be NULL.
The estimated maximum dimension of this cluster's wound or wounds (e.g. length, depth, etc., as applicable), in centimeters.
This column may be NULL, when a dimension is not
recorded or not applicable.
A character, indicating if this cluster's
wound/pathology impairs the individual's locomotion. Legal
values are Y,
N, or
U, meaning "Yes",
"No", and "Unknown" (or Uncertain, or Unspecified),
respectively.
This column may not be NULL.
A character, indicating if signs of infection
(e.g. oozing, stiffness, redness) were observed. Legal
values are Y,
N, or
U, meaning "Yes",
"No", and "Unknown" (or Uncertain, or Unspecified),
respectively.
This column may not be NULL.
Comments or descriptive notes about this wound/pathology.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for each instance where an observer provides an update on a report. These updates discuss how the wounds/pathologies have healed (or possibly how they haven't healed), so these updates are generally referred to as "heal updates".
Heal updates may be very specific, referring to a
particular body part (a WP_AFFECTEDPARTS
row). They may be a bit more vague and refer only to a
particular wound/pathology (a WP_DETAILS
row). They may even be so vague that the identity of the
report (a WP_REPORTS row) being updated is
the only "known" datum. To flexibly accommodate this
variation, this table includes the WPRId, WPDId, and WPAId columns. These columns allow
the recording of the report being updated, the particular
wound/pathology from that report, or the body part affected by
that wound/pathology, respectively. In each row of this table,
one of these columns (WPRId,
WPDId, and WPAId) must not be NULL, and the
others must be NULL.
The Date in this table must be on or after the associated report's Date.
A heal update may indicate that an individual is
missing, or presumed dead. For this reason, the Date may be after the individual's
Statdate. However, the system will
send a warning when the Date
is more than
90 days after
the individual's Statdate.
When wounds/pathologies are especially severe or
life-changing, heal updates may continue for years after the
related Date. However, these are
rare. The system will return a warning when a Date is more than
365 days after its
related Date.
Tip
Use the WP_HEALS view instead of this table. It presents the data in a format more hospitable for humans to read, and performs the somewhat-tricky task of joining the different ID columns to their respective wound/pathology tables.
A unique identifier for the row. This is an automatically generated sequential number that uniquely identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The WP_REPORTS.WPRId of the related wound/pathology report, if a more specific indicator is not known.
The WPDId of the related wound/pathology cluster, if known and if the related body part being updated is not known.
The WPAId of the related body part, if known.
The WP_HEALSTATUSES.HealStatus indicating how well the related wound(s)/pathology(ies) have healed.
This column may not be NULL.
Textual notes about the healing (or lack thereof) in this update.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table records the observers of the wound/pathology reports, one row for each observer. When a report has multiple observers, each of them is recorded in this table in a separate row.
Each WPRId-Observer combination must be unique; a report cannot have the same observer more than once.
Tip
Use the WP_REPORTS_OBSERVERS view to insert data into this table. It provides a simple way to determine the appropriate WPRId value to use, and for a human data enterer to provide multiple observers in a single row.
Tip
Use the WP_HEALS or WOUNDSPATHOLOGIES views to select data from this table. These views include related data from the other wound/pathology tables, respectively with and without related healing updates.
A unique identifier for the row. This is an automatically generated sequential number that uniquely identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The WP_REPORTS.WPRId of the related report.
This column may not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
Records each distinct report of wounds/pathologies for an individual. When a wound or pathology is first seen, field observers usually report it with a specialized form that helps systematically report various pieces of pertinent data. These data may include but are not limited to which kind(s) of wound/pathology was observed, which body part(s) was affected, wound size (if applicable), and updates on later dates describing how well the individual has healed. These reports may describe something small like a single scrape or limp, or something larger like a set of bleeding wounds on several body parts. This table contains one row for each of these wound/pathology "reports".
It is difficult to provide a precise definition of a "report" in this sense. The aforementioned specialized forms are not always used, so a "report" does not always refer to these forms. Some wounds may be especially serious or life-altering, and pathologies may be chronic or reoccuring, meaning that some wounds/pathologies may recur throughout life in several reports. Because of this, each "report" is not necessarily a distinct "instance" of wound/pathology. Frankly, the distinction between "reports" made in this table is mostly artificial, useful for bookkeeping but lacking biological relevance. In general, each "report" is a discrete observation of wounds/pathologies for an individual on a specific date. A "report" may be an elaborate form, a brief note, or something in between.
Each combination of Sname,
Date, and non-NULL Time must be unique; an individual can
have multiple reports on the same date, but not at the same
time.
The Date must be between the individual's Entrydate and Statdate, inclusive; the individual must be alive and in the study population when the report was created. The system will return a warning if the Date is before theindividual's LatestBirth.
The Grp indicates the group written on the form by the observer. For a variety of reasons (e.g. immigrations, group fissions/fusions), the Grp column may be different from the individual's Grp on this Date. Because of this, validation of the Grp column is limited: the Date must be on or after the group's Start date. The system will return a warning when a report occurs after its Grp has ceased to exist; that is, the system will return a warning when the report's Date is after the group's Cease_To_Exist.
The system will return a warning for any WP_REPORTS rows that do not have at least one related row in WP_DETAILS.
Tip
When adding new data to this table, use the WP_REPORTS_OBSERVERS view. It simplifies the process of adding multiple observers to WP_OBSERVERS.
Tip
Use the WP_HEALS or WOUNDSPATHOLOGIES views to select data from this table. These views include related data from the other wound/pathology tables, respectively with and without related healing updates.
A unique identifier for this report. This is an automatically generated sequential number that uniquely identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
A unique identifier for this report, generated by data management.
This column may not be NULL.
The date that the wounds/pathologies in this report were first observed.
This column may not be NULL.
The time that the wounds/pathologies in this report were first observed, if known.
This column may be NULL, when the time is
unknown.
The BIOGRAPH.Sname of the individual whose wounds/pathologies are described in this report.
This column may not be NULL.
The GROUPS.Gid of the group in which the individual was located when the wounds/pathologies were recorded, according to the observer(s).
This column may not be NULL.
Comments or descriptive notes about the wounds/pathologies from the observers on initial observation.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
Records of the initiation and cessation of continuous periods of observation during which all of a female's cycling events are presumed, for the purpose of analysis, to have been observed. This table contains one row for each female for each initiation or cessation of a continuous period of observation.
A female is considered to be under continuous observation when all of her sexual cycle transition events -- Mdates, Tdates, and Ddates -- are observed or clearly implied by observational data.[56] When CYCGAPS contains a record of observation cessation this is an indication that some of a female's sexual cycle events have gone unrecorded. For this reason when the interval enclosed by a Mdate, Tdate, Ddate sequence contains CYCGAPS rows indicating interruption of observation, the sexual cycle transition dates to either side of the interruption must be in different sexual cycles. For further information on this and other ways CYCGAPS interacts with the rest of Babase, see the documentation on the CYCLES, CYCPOINTS, PREGS, and SEXSKINS tables.
The presumption is that females are under continuous
observation -- females with no CYCGAPS are presumed to be
under continuous observation. Consequently a female's
earliest CYCGAPS Code must be
E (End), denoting the end of a
period of observation.
A female may not have two “start of
observation” (Code S)
without an intervening “end of observation” (Code
E), or vice versa. Otherwise
there would be starts without ends or ends without starts.
Single day observation rows ("points", Code
P) may only occur between an
end of observation/start of observation pair of rows. There
must be a 1-day interval between a female's CYCGAPS rows, with
the single exception that an end of observation may be dated
the day after a start of observation. Otherwise the same
pattern of observation could be recorded using fewer
rows.
Rows with a Code value of
S (Start) or
P (single Point), that mark the
beginning of observational periods or that represent isolated
single days of observation, must have a value in the State
column. All other rows, those with a code of
E (End) that represent the end of
an observational period, must have no value (NULL) in the
State column. When a State value is present, it must
correspond to the sexual cycle transition information on CYCPOINTS. For further information regarding
required correspondences between CYCGAPS and CYCPOINTS, and how changes in CYCPOINTS can automatically change CYCGAPS with a
Code of S, see the
CYCPOINTS documentation below.
Note
To simplify updates to this table, all of the above conditions are validated on transaction commit.
Warning
Any changes to the Date or
Code — including
UPDATE and all INSERT
and DELETE commands — cause
cascading updates to the CYCGAPDAYS table
upon transaction commit. However, the
validation for several other tables — especially CYCPOINTS — depends on the accuracy of
CYCGAPDAYS. As a result, transactions
involving simultaneous updates to both CYCGAPS and CYCPOINTS may result in spurious data, because
validation on the latter may not be reliable. Therefore,
when making changes for a given individual to both CYCGAPS
and CYCPOINTS, don't do them in the same
transaction. Specifically, CYCGAPS inserts, updates, or
deletes should be performed in a transaction where no other
tables are affected[57][58].
Only females may have CYCGAPS rows.
This table is used in the construction of the sexual cycle day-by-day tables. It also affects the determination of which sexual cycle events (CYCPOINTS) are part of a single sexual cycle (CYCLES), the construction of automatic Mdates, and the validation of sexual cycles with respect to pregnancies.
Caution
The State value is ignored in all a female's CYCGAPS rows with Dates on or before the female's Matured, excepting the row with the latest date, as the sexual cycle day-by-day tables contain no rows before the date of sexual maturity.
The combination of Sname and Date is unique.
All rows must be while the individual is alive. That is, the Date must be on or after the individual's Birth and on or before her Statdate.
The short name of the female. This column should
contain the Sname of a female
in BIOGRAPH. This column may not be
NULL.
To simplify the database code, this value may not be changed.
What kind of endpoint the date records. Legal values are:
| Code | Mnemonic | Definition |
|---|---|---|
S | Start | the date is the start of a period of observation |
E | End | the date is the end of a period of observation |
P | Point | the date is an isolated observation that belongs with no other observations, it is both a start and an end of an observational period |
The state of the female's sexual cycle on the given date. Valid values are:
| Code | Mnemonic | Definition |
|---|---|---|
M | menses | follicular -- Mdate (inclusive) to Tdate (exclusive) |
S | swelling | follicular -- Tdate (inclusive) to 5 days prior to Ddate (exclusive) |
O | ovulating | 5 days prior to Ddate (inclusive) to Ddate (exclusive) |
D | deturgesence | luteal -- Ddate (inclusive) to Mdate (exclusive) |
P | pregnant | Ddate (exclusive) to birth (exclusive) |
L | lactating | birth (inclusive) to Tdate (exclusive) |
Must not be NULL when Code is
S or
P, must be NULL when code
is E. See discussion in the
table description above.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table records information on the sexual cycle of the females, one row per female per cycle.
Tip
The user may find it easiest to understand the function of this table by considering the CYCPOINTS_CYCLES view, which joins the CYCLES and CYCPOINTS tables.
Rows in this table depend upon rows in CYCPOINTS: Babase automatically manages the creation and destruction of CYCLES on the basis of the the sexual cycle transition events recorded in CYCPOINTS. The fundamental sexual cycle is a Mdate, Tdate, Ddate sequence. The Babase system automatically creates one row in CYCLES for every Mdate, Tdate, Ddate series in CYCPOINTS. Similarly, if an Mdate, Tdate, Ddate series is removed from CYCPOINTS, the corresponding row is removed from CYCLES. However, the rules Babase uses when automatically creating, destroying, or updating CYCLES are complicated by menarche, death, and gaps in observation.
In some cases there are turgescences of small size and short duration that typically occur during a pregnancy, prior to maturity, after a span of time spent in postpartum amenorrhea, or during a time of stress. These brief turgescent periods are not recorded as cycles because they were deemed too brief or small to be biologically functional.
Tip
If it ever becomes desirable to know when these brief turgescences occurred, this information should be recoverable from the SEXSKINS table, where the actual size of a female's turgescence is recorded.
Caution
CYCLES is special in that some of its data are automatically maintained by the system. The columns Seq and Series are updated automatically. For further information see the documentation that follows, and each column's documentation.
Tip
CYCLES rows should always have related CYCPOINTS rows[59], but as a practical matter it is necessary to create the CYCLES row before creating the related CYCPOINTS rows. This requires noting the Cid of the new cycles row so that it can be referenced in the new CYCPOINTS rows. Rather than do this by hand the CYCPOINTS_CYCLES view can be used. This allows a Sname to be specified with each new CYCPOINTS row and leaves it up to the system to either find or create an appropriate CYCLES row.
The system will report as an error those rows on CYCLES
with no related CYCPOINTS rows[60]. CYCLES with no related CYCPOINTS must have a NULL Seq.
The aggregation of CYCPOINTS rows into cycles is automatically managed by Babase. The determination is based on the order in time of a female's CYCPOINTS rows and the information on gaps in observation present in CYCGAPS. The transition events recorded in CYCPOINTS are collected into sexual cycles, each cycle having (at most) an onset of menses date (Mdate), an onset of turgesence date (Tdate), and an onset of deturgesence date (Ddate), appearing in the order given here when ordered by date, and with none of the female's other Mdate, Tdate, or Ddate CYCPOINTS rows on the interval. Some sexual cycles may lack one or more of the transition events. This may occur for biological reasons — there must not be a resumption of menses date (Mdate) in an individual's first adolescent cycle, nor in the firct cycle after a pregnancy — or simply because there are no data available to identify the date(s). In the latter case, CYCGAPS should be updated with a record of the gap in observation and the respective row is omitted from CYCPOINTS.
Part of Babase's automatic management of cycles is the
management of cycle sequence numbers. Babase assigns a
sequence number (Seq) to each of a
female's cycles, beginning with 1 at
menarche and counting up.
As a consequence of the numbering scheme, the sexual cycle
with a sequence (Seq) of
1 must not have an onset of menses date
(Mdate).
Gaps in periods of continuous observation (CYCGAPS) impact Babase's determination of what constitutes a cycle. The presence of a gap in observation forces a change in cycle. (However, gaps in observation, missing cycles, do not cause gaps in the sequence numbering.) The introduction or removal of a gap, or for that matter the addition or removal of new CYCPOINTS rows, can result in the split of an existing cycle into two -- the creation of a new CYCLES row --, or the merging of two previously distinct cycles into one -- the destruction of an existing CYCLES row. When this occurs the later CYCPOINTS rows retain their Cid, it is the earlier CYCPOINTS rows that change their Cid and “move” between cycles.[61][62][63]
The sexual cycles themselves are aggregated
into periods of continuous observation, termed series,
indicated by the assignment of a Series number to each CYCLES row. The aggregation of a female's sexual
cycles into a series is also automatically managed by Babase,
based on the information in CYCGAPS.
Although series are computed based on CYCGAPS, the series value aggregates and numbers
sexual Mdates, Tdates, and Ddates, not periods of observation.
A consequence is that some periods of observation may not have
an associated Series number. Some
observational periods may occur before the female's sexual
maturity date or before any recorded sexual cycle transition
events (CYCPOINTS). An individual's first
period of continuous observation containing Mdates,
Tdates, or Ddates has a Series of 1, the second
a Series of 2,
etc.
Aggregating a female's CYCLES rows into a series indicates that the collection of data points is believed to be complete, no unobserved or unrecorded sexual cycle transitions (CYCPOINTS rows) occurred during the time spanned by the series. This allows the Series to be used as the basis for an analysis of sexual cycle transition intervals.
Tip
Those CYCLES with a Series of 1 for
those females that have an O (On)
Mstatus have Seq values that
equal the ordinal numbering of the female's actual cycles,
her first ever cycle having a Seq of 1,
her second a Seq of 2, etc. All other
CYCLES rows have Seq values that are useful for ordering
each female's cycles but not for comparison between
females.
Caution
Because a gap in observation always triggers a change in cycle, and because cycles must be “complete”, i.e. must contain a Mdate, a Tdate, and a Ddate, if there is no gap in observation it is impossible to have a single cycle missing nothing but a Tdate, i.e. it is impossible to have a cycle with a Mdate and a Ddate but no Tdate. If necessary, an estimated Tdate may be entered to work around this limitation.[64]
The system reports an error when the combination of Sname and Seq is not unique.[65]
A numeric identifier identifying each sexual cycle. It is unique across all cycles of all females.
This column need not be manually specified when the row is created.
The value of this column may not be altered after a row is created.
This column must not be NULL.
The short name of the female. This column must contain the Sname of a female in BIOGRAPH.
The value of this column may not be altered after a row is created.
This column must not be NULL.
The first sexual cycle of a female has a Seq value
of 1, the second a value of
2, etc.
The system will report an error if the Seq does not begin
with 1 or is not contiguous. This
column does not need to be manually maintained.
Caution
There are no gaps in the sequence numbers assigned to a female. Even when records of cycles are missing, the first recorded cycle after the missing period has a sequence one greater than the last recorded cycle before the missing period.
If the user does specify a value for this column the system may recompute and replace the supplied value at any time.
This column may be NULL when the row is first
inserted, so that the system can set the value correctly
when CYCPOINTS are subsequently
inserted, but it may not be changed from a non-NULL
value to NULL.
Number indicating with which series of continuous observation the
transition event belongs. Events that are isolated
observations have a series of their own. As with Seq, the
Series are per-female. Each female begins with a Series
of 1 and is incremented with
each interruption in regular observation. For further
information see the description of the CYCLES table
above.
The system will report an error if the Series does
not begin with 1 or if the Series does
not progress in a contiguous fashion. This column does
not need to be manually maintained.
If the user does specify a value for this column the system may recompute and replace the supplied value at any time.
This column may be NULL when the row is first
inserted, so that the system can set the value correctly
when CYCPOINTS are subsequently
inserted, but it may not be changed from a non-NULL
value to NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table records information on the sexual cycle of the females, one row per female per event.
Tip
The user may find it easiest to understand the function of this table by considering the CYCPOINTS_CYCLES view, which joins the CYCLES and CYCPOINTS tables.
The usual events that mark the transitions of a female
baboon's sexual cycles are onset of menses (Mdate), onset of
turgesence (Tdate), and onset of deturgesence (Ddate). These
different transition event dates are distinguished by Code values of
M,
T, and
D respectively. In
addition to these usual observations of transition states,
CYCPOINTS contains
one other
kind of row,
estimations of when unobserved sexual cycle transitions
occurred; notably the automatically calculated onset of menses
dates but also unobserved onset of deturgesenceses (Ddates)
related to pregnancy conception events[66].
The unusual events that impact female cycling records, notably death and the cessation or initiation of long term observation, are recorded in other tables.
Note
The interval between conception and birth (or fetal death) is the length the pregnancy, by definition, and CYCPOINTS is only place in Babase where conceptions are recorded. For this reason CYCPOINTS includes rows for the Ddate events that begin every pregnancy, including those that record estimated, unobserved, Ddates. It may be that all that is known about a cycle is that a Ddate must have occurred because a pregnancy resulted.
Although Babase requires pregnancies to have a
conception Ddate, and consequently there may be pregnancies
for which an estimated (Source of
E) Ddate must be entered,
there is nothing preventing the user from creating estimated
CYCPOINTS rows for the other Codes.
Caution
CYCPOINTS is special in that some of its data are automatically maintained by the system. The Cid and Source columns can be updated by automatic processes. For further information see the documentation of the CYCLES table and each column's documentation.
The presence of a Ddate row can trigger the automatic generation of a Mdate 13 days later. For further information see the section on Automatic Mdate Generation.
Only Mdates are automatically assigned, and only Mdates
may have a Source of A (Automatic).
Mdates may be manually given a Source of
A, although this may well not be a
good idea as the Automatic Mdate Generation
process may remove the A row at any
point. It is even less of a good idea because automatic
Mdates are not validated, so it is quite
simple to enter an invalid automatic Mdate.
During a period of continuous observation — a series — sexual cycle transition events (CYCPOINTS) should not be missing, except that Mdates cannot be assigned in the case of the first adolescent cycle (at maturity) or at the start of a Resume cycle. An individual's Mdates, Tdates and Ddates should all appear in Mdate-Tdate-Ddate order. The system will report an error if this is not the case.[67] In consequence the combination of Cid and Code must be unique.[68]
Usually a female does not have multiple CYCPOINTS rows for a given date, although there is an exception. A female's onset of menses date (Mdate) may be the same as her onset of turgesence (Tdate) date. Otherwise, none of a female's CYCPOINTS rows may share a date.
Babase allows each sexual cycle transition event to be
associated with 3 dates, the date of record (Date), the
earliest possible date (Edate),
and the latest possible date (Ldate). The earliest (Edate) and latest
(Ldate) possible dates may be NULL. The earliest possible
date (Edate) may not be later than the date of record (Date),
and the latest possible date (Ldate) may not earlier than the
date of record (Date). A female's earliest Tdate may, and
likely will, have an earliest possible date (Edate) assigned
that is before onset of menarche.
A number of constraints on CYCPOINTS involve the
females' sexual maturity dates (MATUREDATES.Matured). When an individual's sexual
maturity date is determined by observation, MATUREDATES.Mstatus
is O (On), her earliest Tdate must be
equal to her sexual maturity date.
Warning
When a female's MATUREDATES.Mstatus is
O (On) her MATUREDATES.Matured is automatically set to her
earliest Tdate. Any error in the Tdate value will be
reflected in the maturity date. This is not true of females
with MATUREDATES.Mstatuses that are not
O. These maturity dates must be
manually maintained.
No date-of-records may occur before a female's maturation date. All of an individual's date-of-record (Date) and late (Ldate) sexual cycle transition date values must be on or after the individual's onset of menarche date (MATUREDATES.Matured). All of an individual's early dates (Edate), Bdates of record (Date), and the first Tdate date-of-record (Date), sexual cycle transition dates must be after the individual's Birth date.
Females with CYCPOINTS rows must have a sexual maturity date. The system will report mature females with no CYCPOINTS rows on or after her maturity date (MATUREDATES.Matured).
All early date (Edate) and date-of-record (Date) values must be on or before the individual's Statdate.
Caution
Even when an individual is dead, late (Ldate) dates may be after the Statdate. This is because death is rarely observed; although the Statdate contains a single date, the uncertainty surrounding the date of death is reflected in the sexual cycle event Ldate.
There are gaps in observation. If the first cycling
event in a series -- the first Mdate, Tdate, or Ddate -- falls
on the day observation resumes then things are pretty simple.
The state of sexual cycling at the time observation resumes,
CYCGAPS.State,
must correspond with the event. For a menses CYCGAPS.State is
M and so forth. The situation
is slightly complicated by the swelling-follicular and
ovulating states. The details are this: If the first
CYCPOINTS row in the series falls on the first day of the
series, the CYCGAPS.State must be
M (Menses, follicular) when
the CYCPOINTS.Code is M (onset of
Menses); CYCGAPS.State must be
D (Deturgesence) when
the CYCPOINTS.Code is D
(onset of Deturgesence); CYCGAPS.State must be
S (Swelling, follicular)
when the CYCPOINTS.Code is T
(onset of Turgesence) and the subsequent Ddate in the series
is more than 5 days after the Tdate or there
is no subsequent Ddate; and CYCGAPS.State must be
O (Ovulating) when the
CYCPOINTS.Code is T (onset of
Turgesence) and the subsequent Ddate in the series is not more
than 5 days after the Tdate.
If the above is not the case, i.e. the first cycling
event in the series falls on the day observation resumes and
CYCPOINTS.Code
is M but the CYCGAPS.State is not,
then the State of the CYCGAPS row is automatically changed to enforce
correspondence between CYCGAPS and
CYCPOINTS.
But what if observation starts and then later the first Mdate, Tdate, or Ddate is observed? What happens (to CYCSTATS) between the start of observation and the first event? That's what CYCGAPS.State is supposed to address and it needs to be set appropriately. This cannot always be done automatically either, although usually it can.
If the first CYCPOINTS row in the series does
not fall on the first day of the series,
the CYCGAPS.State
must be D (Deturgesence)
when the first CYCPOINTS.Code is
M (onset of Menses); the CYCGAPS.State must be
S (Swelling, follicular)
when the CYCPOINTS.Code is
D (onset of Deturgesence)
and the CYCPOINTS.Date is more than 5 days
after the CYCGAPS.Date; and the CYCGAPS.State must be
O (Ovulating) when the
CYCPOINTS.Code is D (onset
of Deturgesence) and the CYCPOINTS.Date is not more than
5 days after the CYCGAPS.Date.
In these cases, as before, the State of the CYCGAPS row is automatically changed to enforce correspondence between CYCGAPS and CYCPOINTS.
The final set of possibilities have to do with Tdates,
which are complicated because they occur at menarche and after
pregnancies, as well as after menses. The system will report
an error if the first CYCPOINTS row in a series does not fall
on the first day of the series and the first CYCPOINTS row is
a Tdate and the CYCGAPS.State is something other than
M (Menses),
P (Pregnant), or
L (Lactating). Because
there are 3 possibilities in this case, the CYCGAPS.State value is
not automatically assigned.
Note
All of the validation and possible updating of the CYCGAPS.State is performed on transaction commit.
Warning
Because deleting CYCPOINTS changes a female's cycling state -- a representation of which Babase keeps in the sexual cycle day-by-day tables -- but not the interval of time during which she was under observation (CYCGAPS), removing Mdates, Tdates, or Ddates from CYCPOINTS at the beginning of a series can, possibly, leave the beginning of the series either in an incorrect state or the correct state for an overly long period of time. This can be equally true when the dates of the first CYCPOINTS in a series are changed. Removing all the CYCPOINTS Mdate, Tdate, and Ddate rows from a series will leave the entire observational period in the State specified by the CYCGAPS row that denotes the start of the observational period. This may or may not be correct, especially when the CYCGAPS.State was automatically changed due to the insertion or deletion of CYCPOINTS rows.
When deleting all sexual cycle transition CYCPOINTS rows from an observational period it is best to delete them all in a single transaction, or to delete later rows before earlier rows. Deleting CYCPOINTS rows from the beginning of the observational period changes the CYCGAPS.State value marking the start of the observational period.
CYCPOINTS rows must not fall in an
interval of no observation, excepting estimated (Source is
E) Ddates (Code
D) that are also conception
events. (See PREGS.Conceive.) None of the different kinds of
date values -- early (Edate), date-of-record (Date), or late
(Ldate) -- of the individual's CYCPOINTS rows may be in an
interval during which the individual is not under observation
--
may fall on a date on which the individual has a row in CYCGAPDAYS.
The system will allow but report as an error CYCPOINTS rows
with a Source of E and a Code
of D that are not
referenced in PREGS.Conceive.[69]
Caution
CYCPOINTS and CYCLES are intimately related. Be sure to read and understand the CYCLES documentation.
Once a row is created it must remain associated with the same female -- any re-assignment of Cid must retain the association between the CYCPOINTS row and the old Cid's female.
Note
There are plans afoot to automatically fill in the early and late dates. The early dates would include the day after the immediately prior census date, the late date would be the day of the immediately following census date. There must also be a mechanism for manually overriding the automatic dates.
Warning
When making changes to data for individuals with observation gaps, avoid updating this table in a transaction that also makes changes to CYCGAPS. See above for more information.
A numeric identifier unique to each row. This is
used to reference the sexual cycle transition elsewhere in
the database. This column may not be NULL.
This column need not be manually assigned when the row is created. It may not be changed.
A numeric identifier identifying each sexual cycle. It is unique across all cycles of all females, but shared by all CYCPOINTS rows comprising a cycle -- a Mdate, Tdate, Ddate sequence -- of a female.[70]
This column need not be manually specified when the row is created using the CYCPOINTS_CYCLES view. If it is not specified, the system will determine with which cycle the row should be associated and assign the correct Cid. Should the system find that the sexual cycle transition date belongs in a new cycle, it will make and assign a new Cid.[71] If the column is specified the system does the same work, but when it is appropriate to create a new cycle the supplied value is used.
As the system does the same amount of work whether or not the user specifies a value, the only utility in specifying a value is to manually assign a specific Cid to a new sexual cycle which Babase would otherwise automatically create.
Tip
When sexual cycle transition dates are incorrectly aggregated into sexual cycles, i.e. when the Cid is wrong, it is probably because the record of when the female was under observation — the data on the CYCGAPS table — is incorrect. Correcting CYCGAPS may correct the problem.
Caution
The system automatically assigns, or re-assigns, Cid values as CYCPOINTS and, especially, CYCGAPS rows are inserted, deleted, and altered to keep the database in a state consistent with the definition of a sexual cycle. For this reason any particular Cid is not guaranteed to forever identify a particular Sname/Date/Code. Cpids may be used for this purpose, or the data itself. For further information see the CYCLES documentation.
Supplying a NULL value causes the system to
recompute the correct value and use it in place of the
NULL.
The date-of-record of the transition event. See the
Protocol for Data
Management: Amboseli Baboon Project for information regarding the determination
of this date from the field data. This column may not be
NULL.
Earliest possible date of the transition event. This
column may be NULL when there is no need to record a
range of date values.
Latest possible date of the transition event. This
column may be NULL when there is no need to record a
range of date values.
Code indicating from whence the data were
derived. D (Data -- the
default) for observed data. A
(Auto) for automatically inserted rows (see Automatic Mdate Generation). E
(Estimated) for estimated values not to be used in other
computations, such as estimated
D dates entered to
relate mothers and pregnancies.
This column may not be changed after the row is created.
The type of sexual cycle transition:
| Code | Description |
|---|---|
M | onset of Menses, a sexual cycle transition event |
T | onset of Turgesence, a sexual cycle transition event |
D | onset of Deturgesence, a sexual cycle transition event |
This column may not be changed.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for each recorded pregnancy. A pregnancy is defined to be an event occurring to a mother; a single pregnancy could result in more than one fetus. The only time there will not be a related BIOGRAPH row for the zygote(s) is when the pregnancy is still in progress[72], otherwise there will always be a BIOGRAPH row that records the progeny of the pregnancy.
The progeny may not be born before being conceived -- the conception date (Ddate via Conceive) of the pregnancy must not be later than the birth date value (Birth) of the associated BIOGRAPH row, the child. The mother may not resume cycling until after birth -- the birth date value of the associated BIOGRAPH row must not be later than the resumption of cycling date values (Resume).
The sequence of a female's pregnancies when ordered by parity must correspond with the sequence when ordered by conception date.
The sequence number (CYCLES.Seq obtained via CYCPOINTS.Cid) of the sexual cycle event immediately following pregnancy (Resume) must always be exactly one more than the sequence number of the sexual cycle event associated with conception (Conceive). Only one pregnancy is allowed per conception event -- each Conceive value differs from all the others. These rules ensure that the resumption date follows the conception date and that there is no overlap of pregnancy time periods, from conception date to birth date or, if known, resumption of sexual cycling date, among the pregnancies associated with a particular female.[73] The female associated with the conception sexual cycle event (Conceive) must be the same as the female associated with the sexual cycle event immediately following pregnancy (Resume).
There must not be a resumption of menses date (Mdate) in the sexual cycle (CYCPOINTS.Cid) of the Resume cycle.
The pregnancy must terminate in a birth or fetal loss
before the female resumes cycling; the only exception is
cessation of observation as described below. The Resume
column must be NULL until there is a row in BIOGRAPH with a Pid
referring to the pregnancy.
Note
Note that the check for pregnancy termination, as well as the parity sequence checks, are not performed until the database transaction is committed. This allows a pregnancy discovered after subsequent pregnancies are already on-record to be added to the database by making multiple changes within a single database transaction. Inserting the new PREGS row, inserting a BIOGRAPH row for the progeny, and then updating the PREGS.Resume of the new pregnancy within a single transaction allows the referential integrity rules to be satisfied when the transaction commits.
Caution
Babase keeps a record of the reproductive state of mature females in the sexual cycle day-by-day tables. If these tables are to be correct Babase must know when each pregnancy ends (see BIOGRAPH.Birth), and when cycling resumes. When there is no record of the end of a pregnancy or resumption of cycling Babase must know whether this is due to cessation of observation or just cessation of data entry.
Babase cannot detect when the user has failed to enter rows in CYCGAPS when observation of a pregnant female has ceased. However, it will report errors and unusual conditions it can detect.
The system will report a warning: when an ongoing pregnancy exceeds 191 days -- when there are more than 191 days between the conception date (PREGS.Conceive) and the Statdate, and there are no progeny recorded for the pregnancy (in BIOGRAPH.Birth), and when there are no gaps in observation (see CYCGAPS) during the 191 day interval; when it appears that a conception date should be estimated but it is not -- when there is no Tdate in the conception cycle but the conception Ddate[74] is not estimated, and there is no gap in observation between the conception date and all of the female's prior CYCPOINTS rows.
The system will report an error: when a female has
sexual cycles while a pregnancy is ongoing[75] -- when the female has Tdate CYCPOINTS rows that post-date her pregnancy's
Conceive date but pre-date gaps in observation, and the
pregnancy has no (NULL) Resume.[76] A female must not have any CYCPOINTS rows that postdate a pregnancy with a
NULL Resume, unless the first CYCPOINTS
row is a Tdate or unless they postdate a gap in observation
following the pregnancy.
Warning
The Resume column is automatically updated by
Babase. so long as there is no gap in observation (See CYCGAPS) between the conception date and the
Tdate that resumes cycling. It is set to the Tdate
immediately following the conception date. The system will
report an error if there is a gap in the observation of
sexual cycle events (CYCPOINTS and the
Resume column is not NULL.[77]
Tip
The temporary creation of a gap in observation (CYCGAPS) allows a conception-birth-resumption sequence to be inserted into a pre-existing series of sexual cycle events (CYCPOINTS).
The contents of this column uniquely identifies the pregnancy record. The Pid must be the mother's Sname followed by the probable parity. Because the Pid is only used to identify the record, it is not necessary to change the Pid just because the parity of the pregnancy is found to have changed. Once a unique Pid is established, it may not be changed. When retrieving data from this table the safe approach is to assume nothing about the contents of this column except that it will uniquely identify a pregnancy.
Note
The preferred way to obtain the bearer of the pregnancy is to find the female associated with the ovulation by joining PREGS.Conceive with CYCPOINTS.Cpid to find CYCPOINTS.Cid, join that with CYCLES.Cid to find CYCLES.Sname, and then use that value to find the mother's BIOGRAPH row.[78][79]
Warning
The Parity column must always be used to obtain a meaningful parity value. As Pids cannot change, should a pregnancy be missed and correction only entered into Babase after the entry of a subsequent pregnancy, the female's subsequent Pid will forever contain an incorrect parity.[80]
The cardinality of the
pregnancy. 1 for a female's first
pregnancy, 2 for a female's second
pregnancy, and so forth. There must not be
“gaps” in the pregnancies, sequenced by
Parity, of any female. When the first pregnancy is known,
the Parity sequence begins with 1. When
the first pregnancy is not known, the Parity sequence
begins with
101.
The parity of a female's first pregnancy must be specified. This tells the system whether the parity sequence begins with 1 or 101. The system will automatically generate the parity of subsequent pregnancies, when the user does not supply a parity. When the user does specify a parity the system compares the supplied value with the value it computes for the column and and raises an error if the two do not match. As a special exception the parity is allowed to be in the 100s rather than the 1s, although the parity must remain sequential and without gaps when only the 10s and 1's place of the female's pregnancy parities are considered. E.g. the parity sequence may be either 1, 2, 3 or 1, 2, 103 but not 1, 2, 104. The 1 in the 100ths place signals that there has been a period of no observation[81] and a pregnancy may have been missed. When a pregnancy's parity is changed from the 1's (or 10's) to the 100s Babase will update the parity of subsequent pregnancies so that they are also in the 100s. Babase will only allow a change from the 100s to the 1s (or 10s) of the smallest of a female's pregnancy parities that are larger than 100 -- the first pregnancy after a period of no observation. In this case Babase will not change the parity of subsequent pregnancies; this must be done manually, from smallest to largest. Babase will not allow a change from the 100s to the 1s (or 10s) of a female's pregnancy parities that are larger than the smallest parity larger than 100.
Supplying a NULL value for the Parity causes the
system to recompute the correct value, a value one larger
than the parity of the previous pregnancy, and use it in
place of the NULL.
The information related to the Ddate event that initiated the pregnancy. This is the Cpid of a CYCPOINTS row of the mother. The related CYCPOINTS row should record the date of conception and must record a Ddate.
This column must contain a unique datum.
Tip
When the date of conception is estimated because there is no sexual cycle data, the conception date recorded should be 178 days before the recorded birthday.
This column must not be NULL.
The resumption of cycling event (Tdate) of the first
cycle following the pregnancy. This is the Cpid of a row in CYCPOINTS, which must record a Tdate. This
column may be NULL in those cases when resumption of
cycle information is not known. When this column is not
NULL, it should contain a unique datum.
This column may be automatically updated. (See the description of the PREGS table above.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table records textual notes made by field observers about a female's reproductive status. It contains one row for every date on which such a note was recorded, per female.
Most of the data related to a female's reproduction is recorded systematically by observers and stored in the other tables in this section. In addition to those data, observers occasionally record miscellaneous notes or comments related to a female's reproductive state. Those notes are recorded in this table.
This table only records notes about female reproduction; the Sname must be female in BIOGRAPH.
All notes made about a female on a single day are recorded in a single row; every Sname-Date pair must be unique.
Reproductive notes can only be recorded while the female is alive and under observation; the Date must be between the female's Entrydate and Statdate, inclusive.
It is rare but possible for a note to be recorded before a female reaches sexual maturity. The system will return a warning for rows that are before a female's Matured date, or for rows with females who do not appear in MATUREDATES at all.
Usually, if an observer took the time to write a note about a female, then they also will have recorded the size and/or color of her paracollosal skin. The system will return a warning if a female does not have a row in SEXSKINS whose Date matches the note's Date.
Tip
The SEXSKINS_REPRO_NOTES view is useful for simultaneous uploading of data to this table and to SEXSKINS.
A unique identifier for the note. This is an automatically generated sequential number that uniquely identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The text of the note.
This column may not be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table records information on the females' sexskins, including size and/or color. It contains one row for every recorded observation of each female's sexskin.
Babase requires sexskin measurements be associated with sexual cycles (CYCLES) in accordance with the rules described in the Sexual Cycle Determination section.
Caution
Because sexskin measurements must be related to a female's sexual cycle (a CYCLES row), her Mdate, Tdate, and Ddate sexual cycle events (her CYCPOINTS rows) must be updated before sexskin information may be entered.
Tip
Use the CYCLES_SEXSKINS, SEXSKINS_CYCLES, or SEXSKINS_REPRO_NOTES views to maintain this table.
Note
The checks that compare all the sexskins of a particular cycle raise their errors immediately when the error is a result of changes made directly to the SEXSKINS table. But, should an error condition be created as a result of automatic shifting of sexskins between cycles due to changes to the sexual cycle dates (See CYCPOINTS) the errors are not immediately reported.
Tdates normally occur at some point during the
transition from sexskin Size 0 to Size
1, but can
occur during the transition from sexskin Size
0 to Size
5. Measurements
larger than 5 cannot come on or before
the Tdate of the cycle. The system will generate a warning
when there is sexskin measurement larger than
1 before the
Tdate. The Tdate of a cycle must be after the dates of all
the cycle's sexskin measurements of zero that precede the
earliest 1 or greater measurement occurring in the
cycle.
A Ddate occurs when the sexskin begins to deturgesce. The Ddate of a cycle must be after the last measurement before the largest measurement of the cycle.[82] The system will report a warning when Ddates occur after sexskin turgesence has begun to subside -- Ddates after the first measurement following the largest sexskin measurement(s) of the cycle.
Sexskin turgesence normally begins after menses, so
sexskin measurements (the Size)
before the related cycle's Mdate cannot be larger than
0. When the Size is greater than
0 and there is no
Mdate in the sexual cycle to which the SEXSKINS row is
assigned, the system will generate an error unless the sexual
cycle's Tdate falls on the individual's MATUREDATES.Matured
date and the maturity date is an “ON”
date[83], or the cycle is the first after a pregnancy (the
Cid is a PREGS.Resume value), or the cycle's first CYCPOINTS row after a (CYCGAPS)
gap is 30 or fewer days after
that gap's end date. In the latter case the system will
generate a warning. The sexskin
measurement on the Mdate cannot be larger than
1, unless the Mdate is
also a Tdate in which case the measurement cannot be larger
than 5. The system will
generate a warning when the sexskin measurement on the Mdate
is larger than
0.
In constrast with the Size column, the Color column has no rules governing which values are allowed during different stages of a cycle.
Sexskin rows associated with one cycle must not be contemporaneous with Mdates, Tdates, Ddates, or sexskin turgesence observations related to a different cycle. All of the SEXSKINS Date values associated with a particular cycle must be later than the Mdate, Tdate, and Ddate of the previous cycle and earlier than the Mdate, Tdate, and Ddate of the succeeding cycle. There must not be any overlap of the cycles' sexskin measurement dates, over the time period from a cycle's earliest sexskin measurement date to its latest, between the sexskin measurement dates of a female's different cycles.
Sexskin observations cannot occur during gaps in observation. That is, each row's Date cannot be during any of the individual's gap periods in CYCGAPS. However, there is an exception: sexskin observations are allowed on the date of "point" observations in CYCGAPS.
The combination of Sname, from the associated CYCLES row, and Date must be unique.
The combination of Date and Cid must be unique.
Usually the observer records both the size and the color
on a date, but occasionally they might only one record one and
not the other. Because of this, the system allows either of
the Size and Color columns to be NULL, but will also
return a warning in this case. It is an error if both of
those columns are NULL.
A unique integer which identifies the SEXSKINS row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The CYCLES identifier associated with the sexskin measurement. This is a Cid from the CYCPOINTS table. This column can be used to retrieve the Sname of the female that was measured as well as all other data collected on the cycle.
This column is automatically assigned by the system. Although some (arbitrary) cycle must be associated with the SEXSKINS row upon insert in order to relate the row to a female, the system always uses the Sexual Cycle Determination rules to re-assign the row to the appropriate cycle.
This column may not be NULL.
The date of the observation. This date must be after
the individuals Birth date.
The date must not be after the individual's Statdate. This column may not be
NULL.
This column contains a number indicating the size of
the sexskin in a metric with units that are integers, with
the exception that 0.5 value
is allowed, ranging from 0
through 20, inclusive.
This column may be NULL, but only when the Color is not NULL.
A PCSCOLORS.Color code indicating the observed paracallosal skin color.
This column may be NULL, but only when the Size is not NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
One row for every unstructured data collection event recorded during all-occurrences protocols. The ALLMISCS row containing data collected during a particular sample is related to the SAMPLES row representing the sample. Samples do not have a fixed number of related rows on ALLMISCS, any particular sample may have one, none, or many. Further information may be found on SAMPLES.
A variety of ad-libitum data may be collected during sample data collection. Some of these ad-libitum data can be placed in the INTERACT_DATA and POINT_DATA tables, in which case ALLMISCS is not involved. The data that does not conform to the design of INTERACT_DATA and POINT_DATA is kept in the ALLMISC table.
Note
Consortships recorded as ad-libitum data during focal point sampling are not stored on INTERACT_DATA because INTERACT_DATA requires that consortships have a starting and an ending time and data collected during focal point sampling is without duration. Such consortship data are stored as an ALLMISCS row. Babase presumes that all consortships are recorded systematically during the day on paper and entered into Babase and so it is not necessary to attempt to place ad-libitum consortship data recorded during focal sampling into INTERACT_DATA. . Consortship data are collected during focal samples in order to note whether focal animals are engaged in consortships during a particular sample, and not to record the consortship per se.
Note
Mounts involving the focal individual during all-occurrences sampling are recorded both in the focal sample data and on the paper field ad-libitum records. Consequently, to avoid duplicates in INTERACT_DATA, Babase stores the mounts recorded in the focal data in the ALLMISCS table, but not the INTERACT_DATA table. Mounts in the ALLMISCS table are therefore redundant and may be ignored.
Warning
Babase does the same thing with ejaculations recorded in the focal data as it does with mounts: it records them in ALLMISCS rather than INTERACT_DATA. However, the protocol says nothing about ejaculations occurring during all-occurrences sampling. Anyone researching ejaculations will need to investigate this further.
For further information regarding the information collected see the Amboseli Baboon Research Project Monitoring Guide. For further information regarding which ad-libitum data winds up in ALLMISCS see the Protocol for Data Management: Amboseli Baboon Project. For further information on the structure of the ad-libitum text that is eventually stored in ALLMISCS, see the documentation for the focal sampling data collection program, or see the Amboseli Baboon Research Project Monitoring Guide if the focal sampling data were handwritten.
The combination of Sid and Time must be unique.
A unique integer which identifies the ALLMISCS row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The time the ad-libitum data were taken. This column stores the time using a data type having a precision of one second but the precision and accuracy of the data values are dependent upon the focal data collection system's timekeeping, the operator, and the protocol and is surely not one second. Consult the Amboseli Baboon Research Project Monitoring Guide.
The time may not be before
05:00 and may not be after
19:00.
The unstructured ad-libitum information collected.
At present the text in this column actually does have some structure[84] but appears in ALLMISCS because Babase contains no other place suitable for the storage of the data. The text begins with a one letter code followed by a comma. The allowed one letter codes and their meaning are:
CConsortship. This is redundant information. Because consortships happen over time these consortships should always also be independently recorded and therefore independently entered into INTERACT_DATA and PARTS.
UUnknown. This was once reserved for meta-information -- the field data collection team's comments on the process of data collection -- but its meaning has since become confused with the
Ocode.OOther. Other information about the baboons or their environment. Its meaning has become confused with the
Ucode.
For further information see the Amboseli Baboon Research Project Monitoring Guide.
This column may not be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
One row for every MPIS row (multiparty interaction) involving a consortship. This table extends the MPIS table to include information about consortships.[85]
A unique integer which identifies the MPIS row -- the multiparty interaction.
Because the CONSORTS table extends the MPIS table, the two tables have a one-to-one relationship, this value also uniquely identifies the CONSORTS row.
The value of this column may not be changed.
The disputed female. A BIOGRAPH.Sname of a female.
This column may be NULL when the consorted female
is unrecorded.
The male who consorted with the female prior to the multiparty interaction. A BIOGRAPH.Sname of a male.
This column may not be NULL.
The male who consorted with the female after the multiparty interaction. A BIOGRAPH.Sname of a male.
This column may not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains data from focal sampling points during which the observer recorded information about the focal individual's infant, one row for each focal sampling point. Any focal sampling protocol that includes recording this kind of information is almost certainly going to require that the focal individual be a female, hence the "F" in this table's name.
Note
Despite its name, this table does not require that a focal individual be of any particular Sex. Requirements like those are set and enforced by the STYPES.Sex column.
Whether or not a focal sample is allowed to have data in this table is determined by the sample's SAMPLES.SType and that SType's related STYPES.Has_FPoints value. See the STYPES table for more information.
Each FPOINTS row is connected to a POINT_DATA row via the Pntid column. That is, each row in this
table must have exactly one row in POINT_DATA with the same Pntid. The system will report a
warning for those POINT_DATA rows that
belong to a sample whose SType's related Has_FPoints is TRUE but which do not have
a related FPOINTS row. While every FPOINTS row must have a
related row in POINT_DATA, not every POINT_DATA row has a related FPOINTS row.
Tip
Because every FPOINTS row must have a related POINT_DATA row, when entering a point the POINT_DATA row must be entered before the FPOINTS row.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row. Pntid links FPOINTS with POINT_DATA in a one-to-one manner.
This column may not be NULL.
The position of the infant with respect to the focal female. The legal values for this column are defined by the KIDCONTACTS support table.
This column may not be NULL.
The suckling activity of the infant. The legal values for this column are defined by the SUCKLES support table.
This column may not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains a row for every recorded interaction between animals, including all-occurrence data taken during focal point samples but excluding multiparty interactions (MPI_DATA). Each row records when the interaction occurred. Further information on the interaction is stored elsewhere, notably PARTS. Each interaction in INTERACT_DATA is represented as though it occurs between two ordered individuals designated “actor” and “actee” -- thus resulting in two rows in the PARTS table.
Warning
The INTERACT view should always be used in place of this table. (See Views for the rationale.) INTERACT is an extension of this table which may be useful. It is identical to INTERACT_DATA but is extended with alternate representations of dates and times.
Tip
The ACTOR_ACTEES view provides a way to view interactions as a single rows.[86]
The actual Date of an
interaction is usually known. However, in some cases only the
year and month of an interaction were recorded without specifying
the day. The specificity (or lack thereof) of the Date is indicated by the boolean
Exact_Date column. When Exact_Date is FALSE, this
indicates that the year and month of the Date are known, but not the day. In
these cases, the Date must be
the first day of the month.
The Date of the interaction is constrained by various related dates of its participants, as follows:
The Date cannot be before a participant's Entrydate, with one exception. When Exact_Date is
FALSEthe Date can be before the participant's Entrydate, but the month and year of the Date cannot be before those of the participant's Entrydate.A female may not participate in a mount, consortship, or ejaculation interaction before menarche (MATUREDATES.Matured). When Exact_Date is
FALSEthe Date may be before her Matured date, but the month and year of the Date cannot be before those of her Matured date.A male may not participate in a mount, consortship, or ejaculation interaction before 4 years of age[87]. When Exact_Date is
FALSEthe Date may be before he reaches that age, but the month and year of the Date cannot be before the month and year in which he reaches that age.The system will return a warning when the Date is before the LatestBirth of either participant in the interaction. When Exact_Date is
FALSE, the Date may be before the LatestBirth but the month and year of the Date cannot be before the month and year of the LatestBirth.
Many rules surrounding
INTERACT_DATA's values are closely tied to the project's data
collection protocols. There are two sorts of data collected
on behavioral interactions:
all-occurrences data and
ad-libitum data. All occurrences
data are collected only during focal animal samples. They are
data on all the occurrences of a particular behavior or
interaction during a given time interval and/or involving a
participating focal individual.[88] All occurrences data will always have an
INTERACT_DATA.Sid that is not NULL. Ad-libitum data are
data that are collected opportunistically at the will of the
observer; we do not assume that ad lib data capture all the
occurrences of a given behavior. Ad-libitum data, which
generally are not collected as part of focal animal samples,
usually have a NULL Sid value (only those collected during
a focal animal sample have a non-NULL Sid). Some sorts of
interactions are only collected during focal sampling and
not as ad libitum data outside of focal samples. Approach
(ACTS.Class =
P), and request to groom
(ACTS.Class =
R) are these
interactions; they are only collected during all-occurrences
sampling and must have a non-NULL Sid. Although
consortship and mount[89] data are collected as all-occurrences data
during focal point samples, these data are also collected,
simultaneously and in more detail, in ad libitum
notes. Consequently, they appear in Babase as ad libitum
data in INTERACT_DATA, not as all occurrences data, and
consortships (ACTS.Class =
C), mounts (ACTS.Class =
M), and ejaculation (ACTS.Class =
E) rows always have a
NULL Sid.
Tip
An individual's all-occurrences interactions can be distinguished from ad-libitum data by using the Sid column to reference SAMPLES to see if the individual is the focal of an all-occurrences sample. An example is presented in Appendix B.
Note
INTERACT_DATA rows having a related SAMPLES row, having a non-NULL Sid[90], will automatically have an Observer value equal to the value
in the related SAMPLES.Observer column -- the system
automatically synchronizes observer values between related
INTERACT_DATA and SAMPLES rows. Such
automatically assigned values cannot be changed. To change
the observer the SAMPLES.Observer column must be changed.
Caution
Care must be taken when breaking a relationship
between INTERACT_DATA and SAMPLES, when
setting INTERACT_DATA.Sid to
NULL. The automatically assigned INTERACT_DATA.Observer value may no longer be
correct and so may require manual adjustment.
An INTERACT_DATA row with a NULL Sid and a non-NULL Observer cannot be updated with a
non-NULL Sid unless the
Observer value is also set to
NULL -- manually assigning an observer to an ad-lib
interaction precludes relating the interaction to a focal
point sampling period. Setting Observer to NULL when changing
Sid to a non-NULL value
causes the system to automatically assign the correct value to
Observer -- causes the system
to automatically synchronize observers.[91] Likewise, an INTERACT_DATA row with a non-NULL
Sid cannot be inserted
unless the Observer value is
either NULL or matches that of the related SAMPLES.Observer value
-- new focal sample interactions must be consistent with
respect to the observers recorded in the INTERACT_DATA and
SAMPLES tables. When an INTERACT_DATA row
with a non-NULL Sid and a
NULL Observer value is
inserted then the Observer
value is automatically updated with the related SAMPLES.Observer value
-- again, the observer associated with the interaction is
automatically brought into sync with the focal
sample.
INTERACT_DATA encodes interaction time and duration by
storing the start and stop times of the interaction. The
columns Start and Stop are used for this purpose.
Consortships may have a NULL in either the Start or the Stop
time when the respective value is unknown, otherwise the Start
time must precede the Stop time. Ad-libitum sample agonism
and grooming interactions (ACTS.Class values of
A and
G respectively) must have a
NULL in both the Start and Stop columns. All-occurrences
agonism, grooming, approach (ACTS.Class = P),
and request to groom (ACTS.Class =
R) interactions must
have non-NULL Start times that equal Stop times. Start
always equals Stop for mounts (ACTS.Class = M) and
ejaculations (ACTS.Class =
E).
The columns of this table that contain times, Start and
Stop, are stored using a data type that has a precision of 1
second. The Amboseli
Baboon Research Project Monitoring Guide must be consulted regarding
the precision and accuracy of these data. It is expected that
ad-libitum datum is entered with a 1 minute
precision.[92] Consequently the seconds portion of the time
values must always be 0 when Sid is NULL. All-occurrences
interaction data (Sid is not NULL) do contain
seconds.[93]
When more than one observer is with a group at the same time, they are responsible for making sure that each interaction is only recorded in only one notebook, not duplicated across multiple observers' notebooks (see the Amboseli Baboon Research Project Monitoring Guide for more details). For this reason, it should be emphasized that the Observer column only indicates who recorded this row's interaction (when known), not who actually saw it.
The system will report a warning for interactions which occur between individuals who are not in the same group on the date of the interaction.
A positive integer that uniquely identifies the
interaction. This number is assigned by the system. This
column must not be NULL.
The origin of the data. When the interaction data
were collected during all-occurrences sampling this column
holds a SAMPLES.Sid identifying the all-occurrences
sample during which the data were collected, otherwise
this column is NULL.
A code indicating the kind of interaction. The ACTS support table defines the legal values for this column.
Note
Although Act contains ACTS.Act values, it is often the broader ACTS.Class classification that is of interest.
This column may not be NULL.
The time the interaction began or, in the case of all-occurrences data, the time the interaction was recorded in the field.
The data type of this column has a 1 second precision. The precision and accuracy of the data itself is dependent upon the protocol and the operator and is almost surely not 1 second. Consult the Amboseli Baboon Research Project Monitoring Guide.
The time may not be before
05:00 and may not be after
20:00.
This column may be NULL.
The time the interaction stopped or, in the case of all-occurrences data, the time the interaction was recorded in the field.
The data type of this column has a 1 second precision. The precision and accuracy of the data itself is dependent upon the protocol and the operator and is almost surely not 1 second. Consult the Amboseli Baboon Research Project Monitoring Guide.
The time may not be before
05:00 and may not be after
20:00.
This column may be NULL.
A boolean indicating whether or not the observer
recorded the interaction by hand[94]
. This value is TRUE if yes, FALSE if no.
This column may not be NULL.
A boolean indicating whether or not the Date is the specific date of the interaction.
This column defaults to TRUE, and cannot be
NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
One row for each collection of multiparty interactions.
Multiparty interactions are recorded as an ordered series of dyadic interactions. Each complete series has a single MPIS row in the database.
Note
This is a separate data set from the dyadic interactions recorded in INTERACT_DATA and related tables. Interactions appearing there do not appear in the multiparty interaction data, or vice versa.
The date of the multiparty interaction must be between the Entrydate and Statdate, inclusive, of all the participants. The system will return a warning for each participant whose LatestBirth is after the date of the interaction.
The two participants in the dyadic interactions must be different individuals, the two MPI_PARTS.Snames must be different.
The Context column must be NULL when the Context_type
value is N, no
context.
The Context_type column must be C
(Consortship) and the Context column must be NULL when a
related CONSORTS row exists. The system
will generate a warning when the Context_type column is
C and there is no
related CONSORTS row.
A unique integer which identifies the MPIS row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
Multiparty interactions may be categorized by the context in which they occur. This column identifies the context of the multiparty interaction.
The legal values of this column are defined by the
CONTEXT_TYPES support table. This
column may not be NULL.
Unstructured text describing the context in which the multiparty interaction occurred.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The OBSERVERS.Initials of the person who recorded this multiparty interaction.
This column may be NULL, indicating that the
observer is not known or was not recorded.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
Multiparty interactions are recorded as collections of individual dyadic interactions. This table contains one row for every dyadic interaction of a multiparty interaction collection. Each interaction is represented as though it occurs between two ordered individuals designated actor and actee -- these individuals are recorded in the MPI_PARTS table. The dyadic interactions within the collection are time-wise sequenced. Two rows may have the same sequence number (Seq), indicating that the two interactions occurred simultaneously.
Tip
The MPI_EVENTS view provides a convenient way to view multiparty interactions as single rows.
Caution
Babase records little in the way of causality among the various interactions collected together under the multiparty interaction collection umbrella. At the time of this writing the data protocols require that the initial interaction is a kind of agonism or a kind of help request, so that can be considered causal of the remaining interactions. However there is nothing, other than time-wise sequencing, linking particular requests for help with aid supplied. As a result it is impossible, in the general case, to associate help supplied with help requested. For example, an individual may request help twice, from two different individuals, and then receive help from an third individual. The columns recording the results of help requests (Helped and Active) must therefore be used with caution, as must any attempt to correlate the specifics of help given with help requested.
Multiparty interactions which occur simultaneously must have the same MPIAct values.
The system will generate a warning when more than two MPI_DATA rows, sharing a Mpiid, have the same Seq value -- when there are more than two dyadic interactions occurring simultaneously.
The first interaction of a multiparty interaction
(those with a Seq of
1) must be an agonism or a request for
help, the MPIAct value must be that of an MPIACTS row having a Kind value of
A or
R.
The Helped and Active columns are meaningful when the
MPI_DATA row records a request for help.[95] These columns must be NULL when the MPI_DATA
row does not record a request for help, otherwise they must
not be NULL. The system will generate a warning when the
Helped column indicates that no
help was given but there are subsequent interactions which
record help being given (where the MPIAct values have
H MPIACTS.Kind values)
to the individual who requested help. The system will
generate a warning when Active is
TRUE and there are no subsequent
AH interactions where the
help-requestee is the recipient of help in the same
multiparty interaction collection. The system will generate
a warning when Helped is true and
Active is FALSE and there are
no subsequent PH
interactions where the help-requestee is the recipient of
help in the same multiparty interaction collection.
A unique integer which identifies the MPI_DATA row, and thereby the interaction the row records.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
A number identifying the multiparty interaction collection (MPIS) of which the MPI_DATA interaction is a member.
This column cannot be changed and must not be
NULL.
This column records the kind of interaction which took place. The legal values for this column are defined by the MPIACTS support table.
This column may not be NULL.
The first interaction of each multiparty
interaction collection has a Seq value of
1, the second a value of
2, etc. The system will report an
error if the Seq does not begin with 1 or is not
contiguous.
Note
The Seq values need not be unique, per Mpiid. Duplicate sequence numbers are used to indicate simultaneous interactions, as would happen if, e.g., 2 individuals aggressed against 1.
This column may not be NULL.
This column indicates whether help was given, by the
individual from whom help was requested, in response to a
request for help. Helped must be FALSE when help was
requested from an unknown individual.[96]This column contains meaningful information
only for those MPI_DATA rows which record requests for
help. (See above.)
This column is TRUE when help was given and
FALSE when no help was forthcoming.
This column may be NULL.
This column indicates whether help given was active or passive. It contains meaningful information only for those MPI_DATA rows which record requests for help. (See above.)
This column is TRUE when the help supplied was
active and FALSE when either the help supplied was
passive or when no help was supplied. This column is
NULL when the MPIAct value represents an action other
than a request for help.
Caution
When looking for help requests that received passive help always check the Helped value to be sure that help was actually received.
This column may be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains records of participants in the interactions which make up a multiparty interaction collection (MPIS). Each interaction is represented as though it occurs between two individuals designated actor and actee. Interactions between multiple individuals are broken down into interactions between pairs according to rules described in the protocols. Therefore, this table should contain two rows for every record of an interaction (for every row in MPI_DATA), one row to record the actor, and one to record the actee. Rules for classifying individuals as actor or actee are documented below in the description of the Role column.
Tip
The MPI_EVENTS view provides a convenient way to view multiparty interactions as single rows.
Warning
Every MPI_DATA row should be related to exactly two MPI_PARTS rows, otherwise it is an error. However, the system allows this condition to exist. It is presumed that such an error condition will exist for only as long as it takes to enter a complete set of data. The system will report those cases where there are not exactly two MPI_PARTS rows for every MPI_DATA row.
Caution
The data integrity rules require that the MPI_DATA row be entered before the 2 MPI_PARTS rows.
Either the Sname or the
Unksname column must be NULL,
but not both.
The actor and the actee of an interaction, when specified as Snames, must not be the same individual.
A unique integer which identifies the MPI_PARTS row, and thereby the participant in the interaction the row records.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
Multiparty interaction identifier. This column holds
the Mpiid value of the row on
the MPI_DATA table containing further
information on the interaction in which the animal is a
participant. It can be used to retrieve the other
information recorded on the multiparty interaction. There
must be a row in MPI_DATA with an Mpiid of this value. This column
cannot be changed and may not be NULL.
A three-letter code (an id) that uniquely identifies a particular animal (an Sname) in BIOGRAPH. This code can be used to retrieve information, such as the maternal group of the animal, from BIOGRAPH or other places where the animal's three-letter code appears.
This column must not be NULL when the
participating individual is precisely identified and
NULL otherwise.
The nature of the problem when one of the participants in the interaction cannot be precisely identified. The legal values of this column are defined by the PARTUNKS support table.
This column must be NULL when the participating
individual is precisely identified and not NULL
otherwise.
This column designates whether the row records the actor or the actee of the interaction. The two possible values are:
| Code | Mnemonic | Definition |
|---|---|---|
R | Actor | The actor is usually the one performing the act. For the agonism data, the individual that is the winner (does not perform a submissive behavior) is the actor. For help requests, the individual that is requesting the help is the actor. For help supplied, the individual supplying the help is the actor. For grooming data, the individual that is grooming is the actor. And so forth. |
E | Actee | The actee is usually the one that is the recipient of another animal's attentions. For the agonism data, the individual that is the loser (performing a submissive behavior) is the actee. For help requests, the individual of whom help is requested is the actor. For help supplied, the individual to whom the help is supplied is the actor. For grooming data, the individual that is groomed is the actee. And so forth. |
This column may not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains records of the participants in observed interactions between animals. Each row in the table records a participant. Each interaction is represented as though it occurs between two individuals designated actor and actee. Interactions between multiple individuals are broken down into interactions between pairs according to rules described in the protocols. Therefore, this table should contain two rows for every record of an interaction (for every row in INTERACT_DATA), one row to record the actor, and one to record the actee. Rules for classifying individuals as actor or actee are documented below in the description of the Role column.
Caution
Every INTERACT_DATA row must be
related to exactly 2 PARTS rows, excepting
those INTERACT_DATA rows that are
associated with ad-lib focal point sampling -- those that
have non-NULL Sid values.
Ad-lib interactions collected during focal point sampling
are allowed to have only one participant, but only when that
participant is the focal individual. So that data can be
entered the system allows these error conditions to exist
while a transaction is in progress. These
conditions are validated on transaction
commit.
Caution
The data integrity rules require that the INTERACT_DATA row be entered before the 2 PARTS rows.
Tip
The utility in the PARTS table, as opposed to having single rows for interactions as the ACTOR_ACTEES view does, is in writing database queries that search for interaction participants. It is easy to use PARTS to search for a participant without knowing whether the participant is the actor or the actee. The same is not true of the ACTOR_ACTEES view.
Note
It is easy to produce the ACTOR_ACTEES view from INTERACT_DATA and PARTS, but the reverse would not be true. This is why the underlying database representation is as it is and not the reverse.
The actor and the actee of an interaction must not be the same individual.
A unique integer which identifies the PARTS row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
A three-letter code (an id) that uniquely identifies
a particular animal (an Sname) in BIOGRAPH. This code can be used
to retrieve information, such as the maternal group of the
animal, from BIOGRAPH or
other places where the animal's three-letter code
appears. This column may not be NULL.
This column designates whether the row records the actor or the actee of the interaction. The two possible values are:
| Code | Mnemonic | Definition |
|---|---|---|
R | Actor | The actor is usually the one performing the act. For grooming data, the individual that is grooming is the actor. For the agonism data, the individual that is the winner (does not perform a submissive behavior) is the actor. For mounts, consortships, and ejaculations, the male is the actor. |
E | Actee | The actee is usually the one that is the recipient of another animal's attentions. For grooming data, the individual that is groomed is the actee. For the agonism data, the individual that is the loser (performing a submissive behavior) is the actee. For mounts, consortships, and ejaculations, the female is recorded as actee. |
This column may not be NULL.
Interaction identifier. This column holds the Iid value of the row on the
INTERACT_DATA table containing further
information on the interaction in which the animal is a
participant. It can be used to retrieve the other
information recorded on the interaction. There must be a
row in INTERACT_DATA
with an Iid of this value. This column may not be
NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
One row for every point observation collected on a focal individual during a sampling interval. When, for whatever reason, there are no point data collected on the focal individual at the turn of the minute, there is no row on POINT_DATA. The “position” of the points within the sample, Min value, may therefore contain “gaps” -- missing numbers. The “missing numbers” are points taken when the focal animal is out of sight or the point was missed for whatever reason. Babase represents the observational period during which a sample is collected as a SAMPLES row.
Warning
Always use the POINTS view in place of this table (see Views for the rationale.) It contains additional computed columns which may be of interest and is guaranteed to remain consistent in future Babase releases.
A POINT_DATA row must contain a Foodcode when the Activity column indicates the focal is
feeding, otherwise Foodcode must be NULL.
Consistency is enforced with respect to time taken to collect the sample and the number of point observations. The Min value must not be larger than the Mins of the corresponding sample.
Validation of the Activity and Posture columns partially depends on the row's related SAMPLES.SType. The STYPES_ACTIVITIES and STYPES_POSTURES tables define which SType values can be used with which Activity and Posture values, respectively.
Changing the Sid risks data integrity issues that are not easily prevented with simple data checks, especially with the calculating of Minsis. Because of this, the Sid can only be changed by an administrator or superuser.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and is used in other tables to refer to particular points.
This column may not be NULL.
The SAMPLES.Sid of the focal sample during which this point was collected.
This column may not be NULL.
The ordinal number of the point within the sample.
The first point in the sample has a Point value of
1, the second a Point value of
2, etc. Note that these numbers need
not be contiguous since some points are
“lost” during data collection. (See
above.)
This column may not be NULL.
The time the point was recorded. This column stores the time using a data type having a precision of one second. The precision and accuracy of the data values are dependent upon the focal data collection system's timekeeping, the operator, and the protocol and is surely not one second. Consult the Amboseli Baboon Research Project Monitoring Guide.[97]
Warning
It is unlikely that the researcher is interested in this data because, as of January 2006, the field protocols require no particular relationship between the time of the point and the time the observer records the data.
The time may not be before
05:00 and may not be after
19:00.
This column may not be NULL.
The ACTIVITIES.Activity of the individual when the point was taken.
Some values from ACTIVITIES may be restricted, based on the sampling protocol. See STYPES_ACTIVITIES for more information.
This column may not be NULL.
The POSTURES.Posture of the individual when the point was taken.
Some values from POSTURES may be restricted, based on the sampling protocol. See STYPES_POSTURES for more information.
This column may not be NULL.
Food item eaten when the point was taken, if any.
NULL when no food items are eaten. The legal values for
this column are determined by the FOODCODES support table.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
The neighbors of the focal individual are recorded during point sampling. NEIGHBORS contains one row for every neighbor recorded during a point data collection event (minute).
When no neighbor is observed for a particular neighbor type (Ncode), no new rows are added to this table. This is different from how an "unknown" neighbor is recorded, as discussed below.
A focal individual's neighbors are not always
recognizable or for some other reason do not always have a row
in BIOGRAPH. For this reason NEIGHBORS
contains two different columns used to identify the neighbor,
Sname and Unksname. The first for recording known
neighboring individuals and the second for recording unknown
neighboring individuals. One and only one of these columns
must contain a value, the other column must then contain
NULL.[98]
The system will report a warning when the neighbor is not in the same group as the focal individual.
The neighbor must be alive and in the study population on the day of the sample (SAMPLES.Date, as discovered via POINT_DATA.Sid) -- the day of the sample may not be before the neighbor's Entrydate, and may not be after the neighbor's Statdate.[99] This means that the demographic information for a particular time interval must be entered into Babase before the sample data for that interval.
The system will report a warning when the related Date is before a neighbor's LatestBirth.
Each point observation (Pntid value) may have at most one NEIGHBORS row of a given neighbor classification (Ncode value.) The combination of Pntid and Ncode must be unique.
The NCODES table places restrictions on which individuals can be neighbors. One effect of this is to limit the order in which NEIGHBORS may be added to and deleted from Babase.
The sample's focal individual (SAMPLES.Sname, as discovered via POINT_DATA.Sid) may not be her own neighbor.
The combination of Pntid and Sname must be unique.
Validation of the Ncode column partially depends on the row's related SAMPLES.SType. The STYPES_NCODES table defines which SType values can be used with which Ncode values.
A unique integer which identifies the NEIGHBORS row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The POINT_DATA.Pntid of the point in which this neighbor was recorded. Further information related to the entire sample must be found by using POINT_DATA.Sid, the sample identifier.
This column may not be NULL.
The BIOGRAPH.Sname of the neighbor.
This column must be NULL when the neighbor is an
unknown individual or otherwise not in BIOGRAPH, i.e. when the Unksname is not NULL.
The NCODES.Ncode describing the kind of neighbor represented in the row.
Some values from NCODES may be restricted, based on the sampling protocol. See STYPES_NCODES for more information.
This column may not be NULL.
The UNKSNAMES.Unksnamenature code recorded when the neighbor cannot be precisely identified[100].
This column must be NULL when the neighboring
individual is precisely identified, i.e. when the Sname is not NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
One row for every continuous period of time during which data are collected at regular intervals on a specific focal individual. Although the field protocols center around collecting data primarily stored in the POINT_DATA, FPOINTS, and NEIGHBORS tables, other information — normally collected ad-libitum during data collection — may be collected as well and are also associated with the specific sample. Further, a sample is allowed to contain no (animal) information.[101] Each SAMPLES row contains the information pertaining to all the data collected during the sample.
The date of the sample must not be before the focal individual's Entrydate, nor after the focal individual's Statdate. Therefore the demographic data pertaining to any particular time period must be entered into Babase before the sample data collected during that time period.
The system will return a warning when the Date is before the focal individual's LatestBirth.
The number of point observations occurring during the sampling interval (Minsis) must be less than or equal to the total number of minutes elapsed (Mins) during the sampling interval.[102]
Other data integrity checks may be performed on a SAMPLES row — and on related rows in POINT_DATA, FPOINTS, and NEIGHBORS — depending on the data collection protocol used in the focal sample. Each sample's protocol is indicated by its SType, and the details of these other data integrity checks are defined in the STYPES table.
The system will report a warning when the group (Grp) of the focal individual, as recorded on SAMPLES, is not the same as the group MEMBERS records for the focal individual on the date of data collection.
One of the participants in all interactions collected during the sample (see INTERACT_DATA.Sid and PARTS) must be the focal individual.
Focal sampling protocols usually designate how many minutes should elapse in each sample, but for various reasons samples collected in the field may last for fewer than the expected number of minutes. Regardless of the expected number of elapsed minutes in a sample, the actual number and the number of those minutes in which a focal "point" was collected are recorded in the Mins and Minsis columns, respectively. Both of these integer columns cannot be less than zero. Their maximum allowed value depends on the row's SType and related STYPES.Max_Points.
The data collected during a focal sample are complex. To assist the observer with recording it all, these data are often though not always collected with an electronic device — e.g. a handheld phone/tablet — and specialized data collection software. This table uses three columns to record details about the hardware and software — or lack thereof — used for data collection: Collection_System, Programid, and Setupid. The Collection_System indicates the hardware used (e.g. "Samsung Tablet B", "Psion unit 6", "Pen and paper"). The Programid indicates the software that the hardware used, and the Setupid indicates any special configuration file(s) that the software used.
Tip
If a focal sampling arrangement has no particular need
for one of these columns — e.g. samples recorded with
a pen and paper likely won't need Programid nor Setupid
— do not set that column to NULL. Collection_System isn't allowed to be
NULL, and both Programid and
Setupid should only be NULL when
their true values are unknown. Instead, add a row to the
column's respective support table that essentially means
"N/A" and use that value in this table.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and is used in other tables to refer to a particular sample.
This column cannot be changed and must not be
NULL.
The GROUPS.Gid of the focal individual's group, recorded at the time of data collection by the observer.
This column may not be NULL.
The STYPES.SType of the data collection protocol used in this focal sample.
This column may not be NULL.
The total number of minutes which actually elapsed while the sample was collected.
This column may not be NULL.
The actual number of point observations (once per minute) recorded during the sample.
Babase maintains this value automatically by counting the number of POINT_DATA rows associated with the sample. If this value is manually set, Babase compares the supplied value with the value it computes and issues an error if the two do not match.
This column may not be NULL and must be less than
or equal to this row's Mins.
The SAMPLES_COLLECTION_SYSTEMS.Collection_System indicating how the sample's data were collected.
This column may not be NULL.
The PROGRAMIDS.Programid of the software ("program") used on this row's device to collect this sample's data.
This column may be NULL, indicating that this
information is unknown.
The SETUPIDS.Setupid representing the configuration ("setup") file(s) used by this row's software to collect the data in this sample.
This column may be NULL, indicating that this
information is unknown.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
ANESTHS contains one row for each time additional sedation is administered to a darted individual. If no additional sedation was administered then this table should not contain rows related to the darting.
Anesthetic cannot be administered to the same individual more than once at any given time -- the combination of Dartid and Antime must be unique.
Anesthetic cannot be administered before the individual is darted -- the Antime value cannot be before the related DARTINGS.Darttime time.
Anesthetic cannot be administered after the individual recovers from the previous dose -- the Antime value cannot be later than 2 hours after the later of the DARTINGS.Darttime time or the previous administration of additional sedation.
Tip
The ANESTH_STATS view aggregates the multiple administrations of anesthetic given during a darting and so provides a convenient way to analyze ANESTHS rows.[104]
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and is used in other tables to refer to a particular administration of extra sedation.
This column is automatically maintained by the
database, cannot be changed, and must not be NULL.
The darting event during which extra sedation was
administered -- a DARTINGS.Dartid value. This column cannot be
changed and may not be NULL.
Anesthetic administered to extend sedation. The legal values for this column are defined by the DRUGS support table.
This column may not be NULL.
The time additional sedation was administered to the darted individual.
The time zone is Nairobi local time.
The precision of this column is 1 minute -- seconds
and fractions thereof must be 0.
This column may be NULL when there is no record of
what time additional sedation was administered.
The amount of anesthetic administered, in CCs.
The maximum allowed is 1.0CC. The minimum is 0. The precision allowed and accuracy are .01CC.
This column may not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
BODYTEMPS contains one row for each body temperature measurement taken of a darted individual.
The temperature cannot be measured before the individual
is darted or before the individual is picked up -- the Bttime
value cannot be before either the related DARTINGS.Pickuptime
time or[105] the Darttime time. The
temperature cannot be taken after the individual has recovered
from sedation - the Bttime value, when non-NULL, cannot be
later than 2 hours after the later of the
DARTINGS.Darttime
time or the last administration of additional sedation, if
any, as recorded in the ANESTHS table. A
non-NULL Bttime value implies that there must be a known
time of anesthetic administration -- either DARTINGS.Darttime or
ANESTHS.Antime
must be non-NULL.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and may be used to refer to a particular body temperature measurement.
This column is automatically maintained by the
database, cannot be changed, and must not be NULL.
The darting event during which the body temperature
measurement was taken -- a DARTINGS.Dartid value. This column cannot be
changed and may not be NULL.
The measured temperature in degrees Celsius to a
precision of 1/10th of a degree. The minimum allowed value
is 25 degrees and the maximum
45 degrees.
This column may not be NULL.
The time the body temperature of the darted individual was taken.
The time zone is Nairobi local time.
The precision of this column is 1 minute -- seconds
and fractions thereof must be 0.
This column may be NULL when there is no record of
when the body temperature measurement was taken.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
CHESTS contains a row for each chest circumference measurement made of a darted individual.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and may be used to refer to a particular chest circumference measurement.
This column is automatically maintained by the
database, cannot be changed, and must not be NULL.
The darting event during which the chest circumference
measurement was taken -- a DARTINGS.Dartid value. This column cannot be
changed and may not be NULL.
The chest circumference measurement, in centimeters,
with a precision of 1/10th of a centimeter. The minimum
value allowed is 25
centimeters. The maximum value allowed is
99.9 centimeters.
Caution
The value contained in this column may have been adjusted for systematic observational bias. See the Chunadjusted column for more information.
This column may not be NULL.
Some measurements were subject to systemic bias when
taken. When this is known to have occurred the original,
biased measurements are recorded in this column. When there
is no known bias this column is NULL.
When non-NULL this column contains the original
chest circumference measurement, in centimeters, with a
precision of 1/10th of a centimeter. The minimum value
allowed is 25 centimeters.
The maximum value allowed is
99.9 centimeters.
A sequence number indicating the order in which the
measurements were taken. The first chest circumference
measurement taken during a darting has a Chseq value of
1, the second a value of
2, etc.
The system automatically re-computes Chseq values to
ensure that they are contiguous and begin with
1. See the Automatic Sequencing section for further
information.
Initials of the person who performed the measurement. The legal values of this column are defined by the OBSERVERS support table.
This column may be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
CROWNRUMPS contains a row for each crown-to-rump measurement made of a darted individual.
Tip
The CROWNRUMP_STATS view aggregates the multiple crown-to-rump measurements taken during a darting and so provides a convenient way to analyze CROWNRUMPS rows.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and may be used to refer to a particular crown-to-rump measurement.
This column is automatically maintained by the
database, cannot be changed, and must not be NULL.
The darting event during which the crown-to-rump
measurement was taken -- a DARTINGS.Dartid value. This column cannot be
changed and may not be NULL.
The crown-to-rump measurement, in centimeters, with a
precision of 1/10th of a centimeter. The minimum value
allowed is 10
centimeters. The maximum value allowed is
99.9 centimeters.
This column may not be NULL.
A sequence number indicating the order in which the
measurements were taken.The first crown-to-rump measurement
taken during a darting has a CRseq value of
1, the second a value of
2, etc.
The system automatically re-computes CRseq values to
ensure that they are contiguous and begin with
1. See the Automatic Sequencing section for further
information.
Initials of the person who performed the measurement. The legal values of this column are defined by the OBSERVERS support table.
This column may be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
DART_SAMPLES contains one row for every sample type collected in each darting.
The combination of Dartid and DS_Type must be unique.
Tip
The DSAMPLES view also shows these data, one line per Dartid. For some users, this may be a more desirable way to look at these data.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and may be used to refer to all the samples of a particular DS_Type collected during a single darting.
This column cannot be changed. This column may not be NULL.
The darting event during which the indicated samples were collected -- a DARTINGS.Dartid value.
This column cannot be changed. This column may not be NULL.
The DART_SAMPLE_TYPES.DS_Type of this sample.
This column cannot be changed. This column may not be NULL.
The number of samples collected of the type given in the DS_Type column.
This column may not be NULL, must be greater than
zero, and must be between the DART_SAMPLES.DS_Type's corresponding DART_SAMPLE_TYPES.Minimum and DART_SAMPLE_TYPES.Maximum values,
inclusive.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
DARTINGS contains one row for every darting of an animal when data was collected.
The combination of Sname and Date must be unique.
The individual must be alive and in the study population when darted -- the Date must be between the individual's Entrydate and Statdate, inclusive. The system will return a warning when the Date is before the individual's LatestBirth.
The system will report a warning for females darted on or after 2006-01-01 for which there is no related DART_SAMPLES row that indicates a vaginal swab collection.
The Downtime value cannot be before the Darttime value and cannot be more than 1 hour after the Darttime value.
The Pickuptime value cannot be before the Downtime value and cannot be more than 90 minutes after the Downtime value. It also[106] cannot be before Darttime and cannot be more than 90 minutes after Darttime. The system will report a warning if the Pickuptime is more than 30 minutes after the Downtime.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and is used in other tables to refer to a particular darting event.
This column is automatically maintained by the
database, cannot be changed, and must not be NULL.
A three-letter code (an id) that uniquely identifies
the darted animal (an Sname) in BIOGRAPH. This code can be used to
retrieve information from BIOGRAPH or other places where the
animal's three-letter code appears. This column may not be
NULL.
The time the individual was darted -- when the dart was fired. The time zone is Nairobi local time.
The time may not be before
05:00 and may not be after
20:00.
The precision of this column is 1 minute -- seconds
and fractions thereof must be 0.
This column may be NULL when the time of darting is
unknown.
The time the darted individual succumbed to the anesthetic. The time zone is Nairobi local time.
The precision of this column is 1 minute -- seconds
and fractions thereof must be 0.
This column may be NULL when the downtime is not
known.
The time that the darting team picked up the anesthetized individual.
The precision of this column is 1 minute -- seconds
and fractions thereof must be 0.
This column may be NULL when the pickup time is not
known.
Anesthetic administered by the dart. The legal values for this column are defined by the DRUGS support table.
This column may not be NULL.
Mass of the darted individual, in kilograms. The precision of this column is 1/10th of a kilogram. The minimum value allowed is 1Kg. The maximum value allowed is 40Kg.
The system will report a warning when this column is
NULL.[107]
Notes regarding the logistics of the darting. Comments about collars, anesthetic, etc. Consult the Amboseli Baboon Research Project Monitoring Guide for further guidance as to usage.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
Comments about the animal's condition, darting circumstances, etc. during darting. Consult the Amboseli Baboon Research Project Monitoring Guide for further guidance as to usage.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
Notes on the crown-to-rump measurements taken, if any.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
Notes on the chest circumference measurements taken, if any.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
Notes on the ulna length measurements taken, if any.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
Notes on the humerus length measurements taken, if any.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
Ad libitum notes taken on the physiological features of the darted individual, if any.
This column may be NULL.[108]. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
Notes on the PCV measurements taken, if any.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
Notes on the body temperature readings taken, if any.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
Notes that accompany any of the different samples recorded in the DART_SAMPLES table, if any.
This column may be NULL.[109]. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
Notes on the teeth, if any observations on the teeth were made.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
Notes on the canines, if any observations on the teeth were made.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
Notes on the testes measurements taken, if any.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
Notes on the parasite counts done, if any.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
DPHYS contains one row for each darting event during which physiological measurements were taken.
Additional physiological measurements are recorded in the PCVS and BODYTEMPS tables.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and may be used to refer to a particular set of physiological measurements taken during a darting.
This column is automatically maintained by the
database, cannot be changed, and must not be NULL.
The darting event during which the set of
physiological measurements were taken -- a DARTINGS.Dartid
value. This column cannot be changed and may not be
NULL.
The pulse of the individual in beats per minute. The
pulse must be greater than
0.
This column may be NULL.
The respiration rate of the individual measured in
counts per minute. The respiration rate must be greater
than 0.
This column may be NULL.
The state of the right inguinal lymph node. The legal values of this column are defined by the LYMPHSTATES support table.
This column may be NULL.
The state of the left inguinal lymph node. The legal values of this column are defined by the LYMPHSTATES support table.
This column may be NULL.
The state of the right axillary lymph node. The legal values of this column are defined by the LYMPHSTATES support table.
This column may be NULL.
The state of the left axillary lymph node. The legal values of this column are defined by the LYMPHSTATES support table.
This column may be NULL.
The state of the right submandibular lymph node. The legal values of this column are defined by the LYMPHSTATES support table.
This column may be NULL.
The state of the left submandibular lymph node. The legal values of this column are defined by the LYMPHSTATES support table.
This column may be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
HUMERUSES contains a row for each humerus length measurement made of a darted individual.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and may be used to refer to a particular humerus length measurement.
This column is automatically maintained by the
database, cannot be changed, and must not be NULL.
The darting event during which the humerus length
measurement was taken -- a DARTINGS.Dartid value. This column cannot be
changed and may not be NULL.
The humerus length measurement, in centimeters, with a
precision of 1/10th of a centimeter. The minimum value
allowed is 10 centimeters.
The maximum value allowed is
35 centimeters.
Caution
The value contained in this column may have been adjusted for systematic observational bias. See the Huunadjusted column for more information.
This column may not be NULL.
Some measurements were subject to systemic bias when
taken. When this is known to have occurred the original,
biased measurements are recorded in this column. When there
is no known bias this column is NULL.
When non-NULL this column contains the original
humerus length measurement, in centimeters, with a precision
of 1/10th of a centimeter. The minimum value allowed is
10 centimeters. The
maximum value allowed is
35 centimeters.
A sequence number indicating the order in which the
measurements were taken. The first humerus length
measurement taken during a darting has a Huseq value of
1, the second a value of
2, etc.
The system automatically re-computes Huseq values to
ensure that they are contiguous and begin with
1. See the Automatic Sequencing section for further
information.
Initials of the person who performed the measurement. The legal values of this column are defined by the OBSERVERS support table.
This column may be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
PCVS contains one row for each PCV (packed cell volume) measurement taken from a darted individual.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and may be used to refer to a particular PCV measurement.
This column is automatically maintained by the
database, cannot be changed, and must not be NULL.
The darting event during which the PCV measurement was
taken -- a DARTINGS.Dartid value. This column cannot be
changed and may not be NULL.
The packed cell volume measurement. This is a
percentage and must be between
1 and
99, inclusive.
This column may not be NULL.
A sequence number indicating the order in which the
PCV measurements were taken. The first PCV measurement
taken during a darting has a PCVseq value of
1, the second a value of
2, etc.
The system automatically re-computes PCVseq values to
ensure that they are contiguous and begin with
1. See the Automatic Sequencing section for further
information.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
TEETH contains one row for every possible tooth site within the mouth on which data was collected for every darting event during which dentition data was collected. There may not be data on each tooth or tooth site. The absence of a row in this table says nothing about the presence or absence of a particular tooth at the time of darting.
When the tooth is missing, the Tstate is
M, the Tcondition value must be NULL. When the
tooth is not missing Teeth-Tcondition must be
non-NULL.
There may be only one tooth in any given tooth site within the mouth, at any one time -- for any given darting there may be at most one row in TEETH for each tooth site (TOOTHSITES).
Note
While rows in this table record tooth presence/absence and condition in separate columns, these data might not be recorded that way in the field. In dartings from 2006-onward, the tooth's presence/absence is recorded in the same place that indicates a tooth has the "erupting" condition. Between this and the fact that it can be difficult for observers to discriminate between partially- and fully-erupted teeth, a tooth that in fact was still erupting might only be recorded as "present". Thus, erupting teeth might appear in this table without a Tcondition indicating it. Teeth that were recorded as "erupting" can safely be assumed to truly be erupting, however.
In other words: in dartings since 2006 (inclusive), there are likely some cases where an erupting tooth was mistakenly recorded only as 'present', and there is no way to identify when this has occurred.
Warning
When inserting a row into TEETH a NULL Tstate value has special meaning.
Inserted rows with a NULL Tstate
value are silently ignored; no such rows are ever
inserted.[110]
The Tstate column cannot be
changed to a NULL.
Note
The DENT_CODES view may be used to maintain the TEETH table. This view may also be useful when querying. It returns a single row with individual columns for every kind of tooth.
The DENT_SITES view provides a way to query TEETH, returning a single row with individual columns for each position in the mouth.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and may be used to refer to a particular tooth (or tooth site when a tooth is missing).
This column is automatically maintained by the
database, cannot be changed, and must not be NULL.
The darting event during which the tooth examinations
were made -- a DARTINGS.Dartid value. This column cannot be
changed and may not be NULL.
The tooth, or tooth site if the tooth is missing. The legal values of this column are defined by the TOOTHCODES support table.
This column may not be NULL.
The degree to which the tooth exists. The legal values of this column are defined by the TSTATES support table.
This column will never contain a NULL. See the warning above for
more information.
A code rating the physical condition of the tooth. The legal values of this column are defined by the TCONDITIONS support table.
This column may be NULL. See TEETH above.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
TESTES_ARC contains one row for every darting event for every recorded measurement of testicle width and length circumference.
Caution
The TESTES_ARC table contains testes measurements of a portion of the testicle circumference. The TESTES_DIAM table contains testes measurements of the diameter. The two tables are otherwise identical in that they have the same structure and have corresponding validation rules.
Note
The “pairing” of the width and length measurements within this table exists to make data storage convenient; no special relationship is implied regarding the order in which the measurements were taken. For example, if there are 3 length measurements taken during a darting and 2 width measurements the width and length measurements may have been taken in either of the following orders, as well as other possible orders not listed here: length1, length2, length3, width1, width2 or length1, width1, length2, width2, length3. In other words the value of the Seq column describes the order in which the length measurements were taken and the order in which width measurements were taken but says nothing about the interspersing of length and width measurements.[111]
Either the width or the length must be specified -- both
Testwidth and Testlength cannot be
NULL in the same row.
There can only be one measurement taken per darting per testicle per measurement sequence number -- Testseq must be unique per Dartid per Testside.
Once a Testwidth value is NULL
all the rows (for the same darting) with higher Testseq values must also
have a NULL Testwidth value. The
same is true of the Testlength
column.[112]
An individual must be male to have a row in this table.
The system will report a warning when individuals have testes length measurements less than 15mm or have testes width measurements less than 10mm.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and may be used to refer to a particular testes measurement.
This column is automatically maintained by the
database, cannot be changed, and must not be NULL.
The darting event during which the testes measurements
were taken -- a DARTINGS.Dartid value. This column cannot be
changed and may not be NULL.
The testicle measured. The legal values are:
| Code | Description |
|---|---|
| L | the left testicle |
| R | the right testicle |
This column may not be NULL.
The testes length measurement, in millimeters, with a
precision of 1/10th of a millimeter. The minimum value
allowed is 15
millimeters. The maximum value allowed is
140 millimeters.
This column may not be NULL.
The testes width measurement, in millimeters, with a
precision of 1/10th of a millimeter. The minimum value
allowed is 10
millimeters. The maximum value allowed is
95 millimeters.
This column may not be NULL.
A sequence number indicating the order in which the
measurements were taken. The first measurement, of each
testicle, taken during a darting has a Testseq value of
1, the second a value of
2, etc.
The system automatically re-computes Testseq values to
ensure that they are contiguous and begin with
1. Note that the TESTES_ARC rows are
sequenced within Dartid within
Testside whereas the other darting
tables are sequenced only within Dartid. See the Automatic Sequencing section for further
information.
TESTES_DIAM contains one row for every darting event for every recorded measurement of testicle width and length diameter.
Caution
The TESTES_ARC table contains testes measurements of a portion of the testicle circumference. The TESTES_DIAM table contains testes measurements of the diameter. The two tables are otherwise identical in that they have the same structure and have corresponding validation rules.
Note
The “pairing” of the width and length measurements within this table exists to make data storage convenient; no special relationship is implied regarding the order in which the measurements were taken. For example, if there are 3 length measurements taken during a darting and 2 width measurements the width and length measurements may have been taken in either of the following orders, as well as other possible orders not listed here: length1, length2, length3, width1, width2 or length1, width1, length2, width2, length3. In other words the value of the Seq column describes the order in which the length measurements were taken and the order in which width measurements were taken but says nothing about the interspersing of length and width measurements.[113]
Either the width or the length must be specified -- both
Testwidth and Testlength cannot be
NULL in the same row.
There can only be one measurement taken per darting per testicle per measurement sequence number -- Testseq must be unique per Dartid per Testside.
Once a Testwidth value is NULL
all the rows (for the same darting) with higher Testseq values must also
have a NULL Testwidth value. The
same is true of the Testlength
column.[114]
An individual must be male to have a row in this table.
The system will report a warning when individuals have testes length measurements less than 40mm or have testes width measurements less than 25mm.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and may be used to refer to a particular testes measurement.
This column is automatically maintained by the
database, cannot be changed, and must not be NULL.
The darting event during which the testes measurements
were taken -- a DARTINGS.Dartid value. This column cannot be
changed and may not be NULL.
The testicle measured. The legal values are:
| Code | Description |
|---|---|
| L | the left testicle |
| R | the right testicle |
This column may not be NULL.
The testes length measurement, in millimeters, with a
precision of 1/10th of a millimeter. The minimum value
allowed is 15
millimeters. The maximum value allowed is
75 millimeters.
This column may not be NULL.
The testes width measurement, in millimeters, with a
precision of 1/10th of a millimeter. The minimum value
allowed is 10
millimeters. The maximum value allowed is
51 millimeters.
This column may not be NULL.
A sequence number indicating the order in which the
measurements were taken. The first measurement, of each
testicle, taken during a darting has a Testseq value of
1, the second a value of
2, etc.
The system automatically re-computes Testseq values to
ensure that they are contiguous and begin with
1. Note that the TESTES_DIAM rows are
sequenced within Dartid within
Testside whereas the other darting
tables are sequenced only within Dartid. See the Automatic Sequencing section for further
information.
TICKS contains one row for every darting event during which data on ticks and other parasites were recorded.
When a specific number could not be arrived at because
there was a large number of parasites or there was some other
reason why the count could not be taken, Tickcount should be left NULL.
The value of the Tickstatus column is constrained based on the Tickcount value. For further information see the documentation of the TICKSTATUSES support table and the meaning of the table's Special Values.
The combination of Dartid, Bodypart, and Tickkind must be unique.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and may be used to refer to a particular tick count.
This column is automatically maintained by the
database, cannot be changed, and must not be NULL.
The darting event during which the tick count was made
-- a DARTINGS.Dartid value. This column cannot be
changed and may not be NULL.
The part of the body examined for ticks or other parasites. The legal values of this column are defined by the BODYPARTS support table.
This column may not be NULL.
The kind of tick or other parasite, or kind of parasite and it's developmental stage, or kind of parasite indicator counted. The legal values of this column are defined by the PARASITES support table.
This column may not be NULL.
The recorded count of ticks, ticks in the indicated
developmental stage, other parasites, or parasite signs.
The minimum value allowed is
0, the maximum is
250.
This column may be NULL when there were too many
parasites to count or the count was not taken for some other
reason.
A status value indicating whether and what sort of tick count was taken. The legal values of this column are from the Tickstatus column of the TICKSTATUSES table. See the documentation of the TICKSTATUSES support table for more information regarding what values may be used under which conditions.
This column may not be NULL.
Notes on the parasite infestation of the indicated body part.
Caution
Notes pertaining to parasites but not specific to the particular body part examined belong in DARTINGS.Ticknotes.
This column may contain NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
ULNAS contains a row for each ulna length measurement made of a darted individual.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and may be used to refer to a particular ulna length measurement.
This column is automatically maintained by the
database, cannot be changed, and must not be NULL.
The darting event during which the ulna length
measurement was taken -- a DARTINGS.Dartid value. This column cannot be
changed and may not be NULL.
The ulna length measurement, in centimeters, with a
precision of 1/10th of a centimeter. The minimum value
allowed is 10 centimeters.
The maximum value allowed is
35 centimeters.
Caution
The value contained in this column may have been adjusted for systematic observational bias. See the Ulunadjusted column for more information.
This column may not be NULL.
Some measurements were subject to systemic bias when
taken. When this is known to have occurred the original,
biased measurements are recorded in this column. When there
is no known bias this column is NULL.
When non-NULL this column contains the original ulna
length measurement, in centimeters, with a precision of
1/10th of a centimeter. The minimum value allowed is
10 centimeters. The
maximum value allowed is
10 centimeters.
A sequence number indicating the order in which the
measurements were taken.The first ulna length measurement
taken during a darting has a Ulseq value of
1, the second a value of
2, etc.
The system automatically re-computes Ulseq values to
ensure that they are contiguous and begin with
1. See the Automatic Sequencing section for further
information.
Initials of the person who performed the measurement. The legal values of this column are defined by the OBSERVERS support table.
This column may be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
VAGINAL_PHS contains a row for each vaginal pH measurement taken on a darted female.
Tip
The VAGINAL_PH_STATS view aggregates the multiple vaginal pH measurements taken during a darting and so provides a convenient way to analyze VAGINAL_PHS rows.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and may be used to refer to a particular vaginal pH measurement.
This column is automatically maintained by the
database, cannot be changed, and must not be NULL.
The DARTINGS.Dartid of the darting during which this vaginal pH measurement was taken.
This column cannot be changed and must not be
NULL.
The vaginal pH measurement, precise to the nearest
0.5. This must be a number between
4.0 and
10.0.
This column may not be NULL.
A sequence number indicating the order in which the
measurements were taken. The first vaginal pH measurement
taken during a darting has a VPseq value of
1, the second a value of
2, etc.
The system automatically re-computes VPseq values to
ensure that they are contiguous and begin with
1. See the Automatic Sequencing section for further
information.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This section contains data about the origin, identity, location, and various other traits about the tissue and nucleic acid samples in the users' inventory. This includes samples currently residing in the users' inventory, as well as older samples that may have previously been in use but have since been sent to others, consumed, discarded, or lost. Because of this, the data in this section serve as both a historical record of all samples that have ever been in the users' possession and an active record of the samples that are currently in the users' possession.
Note
The text in this section uses the terms "nucleic acid" and "nucleic acid sample" interchangeably[115]. At the time of this writing, the system does not attempt to record details at the molecular level, so the reader can be assured that comments about the location, source, etc. of a specific "nucleic acid" should be interpreted as referring to a sample and not a specific molecule.
Note
A careful reader may notice many similarities between the way data about tissue samples and nucleic acid samples are recorded, and may wonder why the two data sets aren't unified into a single table. When the TISSUE_DATA and NUCACID_DATA tables were initially designed they were not so similar, and there were some design-based reasons to keep them separate. As of Babase 5.6 that is no longer true, and with some adjustments those two tables surely could now be combined into a single table. In retrospect, we should have designed them this way from the beginning. Changing them now would require a non-trivial amount of work, so in separate tables they remain.
This table contains one row for every location that may be used to store tissue or nucleic acid samples.
Samples may be stored in varied locations with different organizations/research groups ("institutions"). The Institution column is included to allow easy segregation of locations across these varying locales.
The name of each distinct location is recorded in the Location column. Different organizations have their own conventions about how to organize and name storage locations, so this code may be a very descriptive and specific space ("Shelf 1, Rack 2, Box 3, Position D") or something more general ("PINK BOX").
Each Institution-Location pair must be unique.
To allow the use of nondescriptive
general Location values but retain
the ability to enforce uniqueness of specific ones, the
boolean column Is_Unique is
included. When Is_Unique is
TRUE, the row's LocId may occur
at most once across both the NUCACID_DATA.LocId
and TISSUE_DATA.LocId columns (once total, not once
per table). When FALSE, the LocId may be used any number of times in
either table.
A unique identifier for the location. This is an automatically generated sequential number that uniquely identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be NULL.
The INSTITUTIONS.Institution indicating the organization or research group at which this row's Location exists.
This column may not be NULL.
A boolean indicating whether or not this location at this institution is unique.
This column defaults to TRUE.
This column may not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every quantification of a nucleic acid sample's concentration.
Tip
Use the NUCACID_CONCS view instead of this table. It contains additional related columns which may be useful.
A nucleic acid sample
cannot be quantified before it was created, before the source
tissue sample was collected, nor before the tissue sample's
donor entered the study population (if applicable); the Conc_Date cannot be before the
related NUCACID_DATA.Creation_Date, TISSUE_DATA.Collection_Date, nor the related BIOGRAPH.Entrydate. These dates already have a
required sequence to them — Entrydate <= Collection_Date <= Creation_Date <= Conc_Date — so in many
cases it may be sufficient for the system to only require that
Conc_Date is after the
Creation_Date. However, any of
these date columns can be NULL, so for the sake of
completeness the system separately checks that Conc_Date is greater than each
of them.
Concentrations are recorded in two columns: the Quantity and the Unit. This allows the recording of concentrations using any desired unit.
Tip
To convert a quantity from one unit to another related one (e.g. pg/μL to ng/μL), not pg/μL to μM), use the convert_conc() function.
Due to various quirks of DNA quantification, it is possible for a Quantity to be a negative number. However, this is expected to be rare and in most cases should not happen. The system will return a warning for any row in this table whose Quantity is negative.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The NUCACID_DATA.NAId of the quantified sample.
This column may not be NULL.
The NUCACID_CONC_METHODS.Conc_Method used to quantify this concentration.
This column may not be NULL.
The date that this concentration was quantified.
This column may be NULL, when the date is
unknown.
The second half of the concentration: the unit. This must be a NUCACID_CONC_UNITS.Unit value.
This column may not be NULL..
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every person involved with the creation of a specific nucleic acid sample. When a nucleic acid sample has multiple creators, each of them is recorded here in a separate row.
Most nucleic acid samples are created via "extraction". This table favors using "creation" rather than "extraction", for reasons explained in the discussion of the NUCACID_DATA table.
Each NAId-Creator combination must be unique; a sample cannot have the same creator more than once.
Tip
Use the NUCACIDS view to insert data into this table. It provides a simple way to determine the appropriate NAId value to use, and for a human data enterer to provide multiple creators in a single row.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The NUCACID_DATA.NAId of the related nucleic acid sample.
This column may not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every nucleic acid sample that is or ever has been in the inventory. With a few exceptions, each nucleic acid sample's "source" tissue sample is recorded in the TId column, and is attributed to a specific individual in the UIId column.
Tip
Always use the NUCACIDS view in place of this table. It contains additional related columns which may be of interest.
When the nucleic sample is attributed to a specific
individual and its source tissue is provided, the individual
indicated in this table must be that of the source tissue.
That is, when the UIId and
TId are both non-NULL, the
UIId in this table must be equal to the TId's TISSUE_DATA.UIId.
This table records a nucleid acid sample's current location using the LocId column. Values in this column constrain and are constrained by values in the TISSUE_DATA.LocId column, and may or may not be unique, as discussed in the LOCATIONS table.
The Name_on_Tube column indicates whatever "name" or other identifying information is recorded on the tube. Because of labeling errors or misidentification in the field, this value may not indicate the true identity of the individual(s) from whom this sample came.
Two columns in this table record information related to the sample's creation: Creation_Date and Creation_Method. Also the related table, NUCACID_CREATORS. In laboratory vernacular, the term "extraction" is usually favored over "creation" for most nucleic acid sample types. However, some samples are not "extracted" and are instead generated via a laboratory procedure (e.g. reverse transcription, dilution, PCR amplification, etc.). Because of this, the generic term "creation" is used here.
A sample's Creation_Date cannot be before the source tissue's Collection_Date, nor before the source individual's Entrydate, if any. It may often be redundant to verify that Creation_Date is on or after both dates, but this redundancy is intended, as discussed above.
This table attempts to keep an ongoing record of a
sample's current volume in the Actual_Vol_ul column. It is left to
the user to judge this column's accuracy, which depends
greatly on 1) how diligently the lab personnel keep the data
manager(s) informed of changes, and 2) the amount of time that
has passed since this volume was determined[116]. To assist users in making these judgments, the
date that the Actual_Vol_ul was
last updated is recorded in the Actual_Vol_Date column. A sample's
current volume cannot be recorded without also recording this
date; both of the Actual_Vol_ul
and Actual_Vol_Date columns
must be NULL or both non-NULL.
A sample cannot have its current volume determined before the sample was created; the Actual_Vol_Date must be on or after the sample's Creation_Date.
It is unlikely, though not impossible, that a sample's volume might increase after its creation. The system will report a warning when a sample's Actual_Vol_ul is greater than its Initial_Vol_ul.
A nucleic acid sample might have been created from one or more other nucleic acid samples. When this occurs, the nucleic acid sources are recorded in the NUCACID_SOURCES table.
When a nucleic acid sample has more than one nucleic acid source, the sample might contain samples from more than one source tissue, and/or more than one individual. In either case, the TId and/or UIId columns may not be applicable to the sample and will need to be validated using alternative rules. Two boolean columns — Multi_Indivs and Multi_TIds — are used to tell the system how to validate those columns.
The Multi_Indivs column
is used to indicate whether this sample's sources are from
multiple individuals, regardless of the number of sources the
sample has. When TRUE, the UIId must be NULL. When FALSE,
the UIId must not be
NULL.
The Multi_TIds column is
used to indicate whether this sample's sources are originally
from multiple tissue samples, or TId's. As above, this is mostly
independent from the number of individuals attributed to the
sample. When TRUE, the TId
must be NULL. When FALSE, the TId must not be NULL.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The TISSUE_DATA.TId of the tissue sample from which this nucleic acid sample originated.
This column may be NULL, indicating that the
sample has multiple TId sources.
The UNIQUE_INDIVS.UIId of the individual to whomm this nucleic acid sample is attributed.
This column may be NULL, indicating that this
sample is attributed to more than one individual.
The LOCATIONS.LocId indicating the current locale and location of the nucleic acid sample.
This column may not be NULL.
The name of the source individual, according to the label on the tube.
This column may be NULL, when there is no
identifying information on the tube. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The NUCACID_TYPES.NucAcid_Type of this nucleic acid sample.
This column may not be NULL.
The date that this nucleic acid sample was created. When the process to generate a sample lasts more than one day, this is the date that the procedure was completed.
This column may be NULL, when the creation date is
unknown.
The NUCACID_CREATION_METHODS.Creation_Method describing how this nucleic acid sample was created.
This column may not be NULL.
The sample's volume, in microliters, when it was first created.
This column may be NULL, when the initial volume
is unknown.
The sample's volume, in microliters, as of the Actual_Vol_Date.
This column may be NULL, when users have not
updated the sample's "current" volume or when the sample
has not yet been used.
The date that the Actual_Vol_ul was determined.
This column may be NULL, when users have not
updated the sample's "current" volume or when the sample
has not yet been used.
A boolean, indicating whether or not this sample is attributed to more than one individual.
This column may not be NULL. It is presumed that most nucleic acid
samples will be attributed to a single individual, so this
column's default value is FALSE.
A boolean, indicating whether or not this sample has more than one source tissue.
This column may not be NULL. It is presumed that most nucleic acid
samples will have a single tissue source, so this column's
default value is FALSE.
Comments or miscellaneous information about this nucleic acid sample.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every name or ID used only at a specific institution (an ID that is "local" to that institution) to describe a particular nucleic acid.
Identity of samples is maintained by the system as much as possible, but when working with samples in the laboratory this is often inconvenient or impractical. Different groups and institutions often have their own systems for giving unique names to their samples, and while these names may be useful and meaningful for humans, they are mostly unhelpful from the database's perspective. They're vulnerable to typos, and can be very confusing when a sample is shared between institutions. However, these "local names" remain important for the people who are actually using these samples, so these identifiers are recorded in this table, one per nucleic acid sample, per institution.
Every combination of NAId and Institution must be unique; an NAId cannot go by more than one local name at the same Institution.
Every combination of Institution and LocalId must be unique; the same local name cannot be used at a single Institution more than once.
The NUCACID_DATA.NAId of the nucleic acid sample.
This column may not be NULL.
The INSTITUTIONS.Institution indicating the organization or research group at which this NaId's name is used.
This column may not be NULL.
The local name used for this NAId at this Institution.
This column may not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every nucleic acid sample having another nucleic acid as its source.
Tip
Always use the NUCACID_SOURCES_EXT view in place of this table. It has additional columns which may be of interest.
Often, nucleic acid samples are created through some "extraction" process in which the nucleic acids are purified from a tissue sample (e.g. a blood draw, a buccal swab, etc.) However, there are also numerous different methods by which nucleic acid samples may instead be created from another nucleic acid sample (e.g PCR[117], reverse transcription, dilution, etc.). In addition to recording the identity of the source nucleic acid, this table includes the Relationship column, which indicates the nature of the connection between the row's nucleic acid and its source nucleic acid. This relationship may be simple enough to explain in a single word (e.g. "DILUTION"), or complex enough to require a lengthy explanation. To allow this flexibility, Relationship is not constrained to a set of legal values in a support table.
A nucleic acid sample cannot indicate itself as its source; the NAId and Source_NAId cannot be equal.
A nucleic acid cannot have been created before its source; the related Creation_Date of this NAId must be on or after the Source_NAId's related Creation_Date.
When a sample has a single source tissue, its source
nucleic acid samples must also be from that tissue and only
that tissue. That is, when a sample's NUCACID_DATA.TId is
not NULL, all of its source nucleic acid samples must also
have that same TId.
When a sample is attributed to a single individual, its
source nucleic acid samples must also be from that individual
and only that individual. That is, when a sample's NUCACID_DATA.UIId
is not NULL, all of its source nucleic acid samples must
also have that same UIId.
A single sample can have multiple sources, but each of those multiple sources must be unique. Every NAId-Source_NAId pair must be unique.
The NUCACID_DATA.NAId of the nucleic acid that has another nucleic acid as its source.
This column may not be NULL.
The NUCACID_DATA.NAId of the source nucleic acid.
This column may not be NULL.
A textual description of how this nucleic acid and its source are connected.
This column may not be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every population under observation, and/or from which tissue or nucleic acid samples have been collected.
In this context, the term "population" refers to a particular species at a specific location. "The baboons in the Amboseli basin in Kenya", for example, are a population. "All baboons", or "all wildlife in the Amboseli basin", are not.
In the common vernacular, a population is often referred to only by the name of its site, e.g. "Gombe" when referring to the Gombe chimpanzees. Because of this, the Pop_Name and Site columns may seem redundant, but when setting vernacular aside it should be obvious that these two columns contain objectively different information. In practice, users may elect to enter the same value in both of these columns, but the two columns remain independent of each other.
PopId 1
has special meaning to the system. Data integrity rules for
the UNIQUE_INDIVS table presume that the
population with this PopId is the population whose
individuals are recorded in BIOGRAPH. No
other code should be created to refer to that
population.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The name of the population.
This column may not be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The scientific name of this population's species.
This column may be NULL, when unknown or not
applicable. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The common name of this population's species.
This column may not be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
A code indicating whether or not the population is wild or captive. The legal values are shown below.
POPULATIONS.Wild_Captive Values
WWild.
CCaptive.
UUnknown.
NANot applicable.
This column may not be NULL.
The location of the population.
This column may not be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
Comments or miscellaneous information about this population.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every tissue sample that is or ever has been in the inventory.
Tip
Always use the TISSUES view in place of this table. It contains additional related columns which may be of interest.
This table records a tissue sample's current location using the LocId column. Values in this column constrain and are constrained by values in the NUCACID_DATA.LocId column, and may or may not be unique, as discussed in the LOCATIONS table.
If a sample was collected from an individual in BIOGRAPH — if the related UNIQUE_INDIVS.UIId
has a PopId of
1 — the
sample's Collection_Date must be
on or after that individual's Entrydate. Depending on the sample's
Tissue_Type, the Collection_Date may also be
constrained by the individual's Statdate. See TISSUE_TYPES for more information.
Note
The Collection_Date and Collection_Time columns are misnomers. Some tissue samples were not collected; rather, their origination might better be described as having been "generated", "aliquotted", "pooled", etc.
The system will return a warning if a sample's Collection_Date is after the
individual's Statdate, but only
when the sample's Tissue_Type
indicates that the Collection_Date is not constrained by
the individual's Statdate. That
is, when the related TISSUE_TYPES.Max_After_Statdate is NULL.
From time to time, field observers may mistakenly record the wrong collection date on a tube. To help identify when this has occurred, the system uses the CENSUS table to confirm whether the Collection_Date is a date that the individual was actually observed[118]. The result of that confirmation is indicated in the Collection_Date_Status column.
Note
When a sample's Collection_Date is not a Date on which the individual was recorded present in CENSUS, the Collection_Date is not necessarily "wrong". There are numerous circumstances in which a sample may have been collected without a census being performed.
Tip
Do not assume that the date written on a sample's label will always match the Collection_Date. When data managers determine that the date written on a label is erroneous, they may be able to determine the true date and update the Collection_Date as needed.
A tissue sample might have been created from one or more other tissue samples. When this occurs, the tissue's sources are recorded in the TISSUE_SOURCES table.
When a tissue sample
has more than one tissue source, the sample might contain
samples from more than one individual. In that case, some
individual-specific columns may not be applicable to the
sample and will need to be validated using alternative rules.
The boolean Multi_Indivs column
is used to tell the system how to validate those columns.
When TRUE, this indicates that the tissue's sources are from
multiple individuals, and therefore:
UIId must be
NULL.Misid_Status must be
NULL.Collection_Date_Status is not calculated and is set to
NULL
When Multi_Indivs is
FALSE, this indicates that the sample is only
associated with a single individual, and therefore:
UIId cannot be
NULL.Misid_Status cannot be
NULL.Collection_Date_Status is calculated and will not be
NULL
The system will return a warning if a sample is
indicated as having multiple individuals, but does not have
multiple tissue sources from different individuals. That is,
if a tissue has a TRUE Multi_Indivs but does not have at
least two tissue sources in TISSUE_SOURCES
from at least two different UIIds.
A unique identifier for the tissue sample. This is an automatically generated sequential number that uniquely identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The UNIQUE_INDIVS.UIId of the individual from whom this tissue sample was collected.
This column may be NULL, as described above.
The LOCATIONS.LocId indicating the current locale and location of the sample.
This column may not be NULL.
The name of the individual from whom this tissue sample was collected, according to the label on the tube.
This column may be NULL, when there is no
identifying information on the tube. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The date the sample was collected or originated.
This column may be NULL, when the date is
unknown.
The time the sample was collected or originated.
This column may be NULL, when the time is
unknown.
The STORAGE_MEDIA.Storage_Medium in which the sample is stored.
This column may not be NULL.
The MISID_STATUSES.Misid_Status of this tissue sample.
This column may be NULL, as described above.
A code indicating whether this row's Collection_Date is or isn't plausible according to available CENSUS data. The legal values are:
| Code | Description |
|---|---|
0 | This individual is part of the main population and has a non-"absent" CENSUS row on this Collection_Date, OR this individual is not part of the main population and we have no basis to question the accuracy of this Collection_Date |
1 | This Collection_Date is NULL, OR this
individual is part of the main population and either i)
has no CENSUS rows on this
Collection_Date or ii) has only "absent" censuses on
this Collection_Date |
This column is automatically maintained by the
database and will only be NULL when Multi_Indivs is TRUE. Attempts
to manually populate or update this column are silently
ignored.
A boolean, indicating whether or not this sample is attributed to multiple individuals. The value of this column affects how other columns are validated, as described above.
This column may not be NULL. It is presumed that most samples will not
be attributed to multiple individuals, so the default
value for this column is FALSE.
Comments or miscellaneous information about this tissue sample.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every name or ID used only at a specific institution (an ID that is "local" to that institution) to describe a particular tissue sample.
For more details about the reason for this table and the difference between a "local" name/identifier and an ID generated by the database, see the discussion for the NUCACID_LOCAL_IDS table.
Every combination of TId and Institution must be unique; a TId cannot go by more than one name at the same Institution.
Every combination of Institution and LocalId must be unique; the same local name cannot be used at a single Institution to describe more than one sample.
The INSTITUTIONS.Institution indicating the locale in which this TId's name is used.
This column may not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every tissue sample having another tissue as its source.
Tip
Always use the TISSUE_SOURCES_EXT view in place of this table. It has additional columns which may be of interest.
In addition to recording the identity of the source tissue, this table includes the Relationship column, which indicates the nature of the connection between the row's tissue and its source tissue. This relationship may be simple enough to explain in a single word (e.g. "ALIQUOT"), or complex enough to require a lengthy explanation. To allow this flexibility, Relationship is not constrained to a set of legal values in a support table.
A tissue sample cannot indicate itself as its source; the TId and Source_TId cannot be equal.
A tissue sample cannot have been collected before its source; the related Collection_Date of this TId must be on or after the Source_TId's related Collection_Date.
Depending on the details of the Relationship, a tissue sample and its source may or may not be from a different individual. The system does not require that the related UIId of the TId and that of the Source_TId be equal. However, the system will return a warning when they are not equal.
A single sample can have multiple sources, but each of those multiple sources must be unique. Every TId-Source_TId pair must be unique.
Usually, a sample should not have sources from different
individuals if it is not indicated as coming from multiple
individuals. It is theoretically possible, so the system will
return a warning when this occurs. That is, when a sample
whose TISSUE_DATA.Multi_Indivs is FALSE has multiple
sources in this table with different UIId's or has any sources with a
TRUE Multi_Indivs.
The TISSUE_DATA.TId of the tissue sample that has another tissue sample as its source.
This column may not be NULL.
The TISSUE_DATA.TId of the source tissue sample.
This column may not be NULL.
A textual description of how this tissue sample and its source are connected.
This column may not be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every individual under observation, and every individual from whom tissue or nucleic acid samples have been collected.
In contrast to BIOGRAPH, which records the identities of every individual in the main study population[119], this table also records the identities of all the individuals in other populations from whom there are tissue or nucleic acid samples recorded in the inventory. All individuals in BIOGRAPH are also included in this table, whether or not tissue or nucleic acid samples exist in the inventory. This presents a problem: there are two tables that separately track the identities of all individuals in the main population. To address this, the triggers have been written to ensure that BIOGRAPH retains primary authority over all individuals in the main population.
Management of individuals in the main population is done by BIOGRAPH (see its discussion for more information), so the ability to perform inserts/updates/deletes in this table for those individuals is heavily constrained, as follows:
Inserting rows for individuals in the main population is only allowed for the unknown individual or for individuals in BIOGRAPH who have not yet been added to this table[120].
The unknown individual's row can only be updated or deleted by an administrator.
Deleting rows for individuals in the main population is only allowed for individuals who are no longer in BIOGRAPH[121].
Updating rows for individuals in the main population is only allowed when changing only the Notes column.
Any individual's PopId cannot be updated to add or remove the individual from the main population.
Tip
Do not manually insert or delete rows in this table for individuals in BIOGRAPH. Perform those actions in BIOGRAPH, and the action will automatically be performed in this table, as well. Manual inserts and deletes in this table should only be done for individuals who are not in BIOGRAPH.
The IndivId column is used to record the individual's name or similar ID. Study projects and research institutions each have their own rules of nomenclature for their individuals, so this might be a lengthy name, an abbreviation, a series of numbers, or some mix of these. This value is not unique; the same identifier may be used more than once across different populations. However, per PopId, each IndivId must be unique; a population cannot use the same identifier more than once.
PopId
1 is the
population recorded in BIOGRAPH, so any
row with this PopId (with a
few exceptions, discussed below) must use the individual's
Bioid as its IndivId.
IndivId
UNKNOWN indicates
the unknown individual, and is allowed to have PopId
1 and not be a
Bioid.
A unique identifier for the individual. This is an automatically generated sequential number that uniquely identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The name/identifier for this individual.
This column may not be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The POPULATIONS.PopId of the individual's population.
This column may not be NULL.
Comments or miscellaneous information about this individual.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This section contains timestamped geolocation data on groups, observers, and significant landscape features (groves, waterholes[122], and possibly other temporary or permanent landmarks), either recorded in a “quad” coordinate system or collected from GPS units. SWERB stands for Sleeping grove, Waterhole, End time, Ranging, and Begin time. Typically SWERB data are collected at hourly or half hourly intervals. Supporting information includes the locations of tree groves and waterholes. For more information see the Protocol for Data Management: Amboseli Baboon Project.
The quad coordinate system was devised prior to the incorporation of GPS technology into the data collection protocols. It is based on regular sub-divisions of the landscape into a grid of squares, 170 m per side. There is no altitude information associated with quad coordinate points. The IDs and coordinates of these quads are recorded in QUAD_DATA.
The GPS X and Y coordinates are in the WGS
1984 UTM Zone 37South coordinate
system. The units of these coordinates are meters, as is the
recorded altitude. The recorded precision of the X and Y values
include at most 1 non-zero digit to the right of the decimal
place, but when the coordinates were recorded using another system the
transformation to UTM may yield values with more digits to the
right of the decimal. X and Y coordinates must be on or within
the bounding rectangle having X coordinates between
42300.0 and 651000.0, inclusive, and Y
coordinates between 9497000.0 and 9894500.0,
inclusive. The system will generate a warning when the location
falls outside the bounding rectangle having X coordinates
between 277000.0 and 311100.0, inclusive,
and Y coordinates between 9689200.0 and
9709500.0, inclusive. The accuracy may vary; see the
Protocol for Data
Management: Amboseli Baboon Project for further information on accuracy at various
times. Altitude is in meters. Altitude values must be between
0 and 10000, inclusive. There must
be no (non-zero) digits to the right of the decimal place for
altitude measurements taken before 2004-01-01. After
2004-01-01 one digit may appear to the right of the
decimal place. The system will generate a warning when altitude
values are NULL but X and Y coordinates are non-NULL.
Some devices and data-exporting applications favor longitude and latitude coordinates via the WGS 1984 2D CRS. Because of this, Babase can read coordinates in that system and transform them to WGS 1984 UTM Zone 37South. Regardless of the coordinate system used when the data are inserted, the coordinates are recorded using UTM. That is, "XYLoc" columns in all Babase tables have the PostGIS "geometry" datatype with SRID 32737, that of WGS 1984 UTM Zone 37South.
All PDOP columns must have values between
0 and
50, inclusive, and have one digit
of precision to the right of the decimal. PDOP values are
unit-less and should be multiplied by the specified accuracy in
meters of the GPS unit to produce a 3 dimensional vector, in
meters, representing the possible distance from the true
location.[123]
All accuracy columns are in meters[124] with one digit of precision to the right of the
decimal and must have values between
0 and
15, inclusive.
The kind of reported error is partially determined by
characteristics of the the GPS unit used for data collection.
GPS units which report error as a PDOP reading, those with GPS_UNITS.Errortype
values of PDOP, cannot be related
to rows with non-NULL Accuracy values. GPS units which report
error as an accuracy reading, those with GPS_UNITS.Errortype
values of accuracy, cannot be
related to rows with non-NULL PDOP values. PDOP values must
be NULL for data collected before 1993-09-01 or after
2001-01-31. Accuracy values must be NULL for data
collected before 2001-02-01.[125] The system will report a warning when data
collected with a GPS unit supporting PDOP or accuracy does not
include, respectively, PDOP or accuracy values.
Warning
On 2000-05-02, the United States government ended its use of Selective Availability, a national security measure which intentionally lowered the accuracy of GPS signals. For more information about this, see Selective Availability on GPS.gov. The GPS accuracy indices in Babase (Accuracy and PDOP) do not and cannot account for this inaccuracy, so users should be aware that any GPS data collected through 2000-05-02 are likely less accurate than indicated.
Warning
GPS data between May and August 2019 are unreliable, apparently thanks to some issues with the European Union's Galileo satellites. See the SWERB Notebook for more information and documentation of this issue.
Starting 2004-01-01, GPS data began to be
downloaded directly from the GPS units instead of being
transcribed by hand. One consequence is that starting
2004-01-01 operators entered up to 10
characters of descriptive codes with each GPS waypoint taken.
This information is processed and distributed throughout the
SWERB data but the various Garmincode columns retain the raw
data as entered by the operator.[126] Before 2004-01-01 the Garmincode columns
must contain a NULL. On or after this date the Garmincode
columns must not be NULL, but may be a string 0 characters
long.[127] SWERB_DATA are the exception to
this rule and may always be NULL. Begin and end rows, rows
with a SWERB_DATA.Event values of
B or
E, may have NULL Garmincode columns regardless of date so
that the data entry staff may supply begin and end rows without
X and Y coordinates should the field team forget to record a
begin or end row. Other SWERB_DATA rows are
except from the Garmincode requirement to handle situations,
notably those which involve lone animals, where data was written
manually for some reason.
Before 2004-01-01 the GPS_Datetime columns must be
NULL. The date portion of the GPS_Datetime columns must
correspond to the date related to containing row. The time
portion of the GPS_Datetime column is not validated, although
the time portion of the GPS_Datetime value occasionally serves
as data against which other columns are validated.
Caution
The Garmincode and GPS_Datetime columns may be NULL, without warning, no matter the date. This is to accommodate the manual recording of data taken using GPS units.[128]
Note
Data is validated per-observation team, per-group, per-day. Data upload and maintenance must be done within transactions that produce valid per-observation team, per-group, per-day data sets.
Note that it may be more convenient to use the views that support the SWERB data than to access the raw data.
This table contains one row for every aerial photo used in the specification of map quadrant system used in the early SWERB data.
A unique identifier of the aerial photo. This is an integer greater than or equal to 1. It is used to refer to a particular aerial photo.
This column may not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for each GPS unit which has been used in the field.
Note
In actual fact early records of unit identification may have been lost. In such cases a row in GPS_UNITS represents a number of units having the same capabilities (i.e. of the same make and model).
The date the unit was first used (Start) must be on or before the date the unit was last used (Finish).
The label on the GPS unit, the Label value must be unique within the time period in during which the GPS unit was in use, between the Start and Finish dates, inclusive.
A 2 digit non-negative numeric value that identifies the GPS unit as a distinct object throughout all time.
This column may not be NULL.
A short textual description of the GPS unit. If necessary this may include additional notes on such details as when the unit was used, its purpose, and so forth.
This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character. This column may not be NULL.
The manufacturer of the GPS unit.
This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character. This column may not be NULL.
The model of the GPS unit. This should be sufficiently detailed that the technical specifications of the unit can be found given this information.[129]
This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character. This column may not be NULL.
The type of error the unit reports. This must be one of:
PDOPThe error is supplied as positional dilution of position.
accuracyThe error is in meters.
See the SWERB Data overview for more information.
This column may not be NULL.
The letter code marked on the unit. Note that this information is not enough to uniquely identify the unit because the same letter codes have been used on different units at different times.
This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character. This column may not be NULL.
The date the GPS device entered service. This date
cannot be before 1993-09-01, the date GPS units were
first used. This column may not be NULL.
The date the GPS unit was taken out of service. This
column may be NULL when the unit is still in
service.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
The QUAD_DATA table contains one row for every map quadrant. For more information, see above in the introduction to the SWERB data.
Note
Before these quads were delineated on 1981-11-01, large scale aerial photographs were used to signify location in SWERB data.
Note
The QUADS view can be used to maintain the QUAD_DATA table. This view may also be more useful than the table when querying.
The unique identifier code used to refer to a
particular map quadrant.[130] This column may not be NULL.
XYLoc (X and Y WGS 1984 UTM Zone 37South coordinates)
The X and Y WGS
1984 UTM Zone 37South coordinates of the centroid of
the map quadrant. This column may be NULL.
See the SWERB Data overview for more information.
Code indicating the aerial photo in which the map quadrant is located, if any. Must be a value on the AERIALS table.
This column may be NULL when there is no aerial
photo for the map quadrant.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every uninterrupted bout of group-level observation for which there is SWERB data.
Start and Stop values are automatically assigned
the SWERB_DATA.Time value from the related SWERB_DATA row with an Event value of
B and
E rows, respectively. The
begin and end of the bout of observation is determined by
the begin and end rows entered in the field (or determined by
the data manager).
Start must be NULL or be
after the related SWERB_DEPARTS_DATA.Time, if any.
The Start value records the
start of the day's observation of the group when there exists
a related SWERB_DATA.Event value of
B and that value is the
first for that group/day and there is no earlier SWERB_DATA.Event
E value. Likewise the Stop value records the end of the day's
observation of the group when there exists a related SWERB_DATA.Event
value of E and that value is
the last for that group/day and there is no later SWERB_DATA.Event
B value. The Start time cannot be after the Stop time.
The Btimeest value is only meaningful when
either there is a begin time value or when investigation of
existing records indicates that no record of a begin time on
file -- when either the Start time
value is non-NULL or the Bsource
value is NR. The Etimeest value is only meaningful when
either there is an end time value or when investigation of
existing records indicates that no record of an end time on
file -- when the Stop time value
is not NULL or the Esource value
is NR. When the values in these
columns are meaningful they must contain a non-NULL value,
otherwise they must contain a NULL value.[131]
When the source of the start or stop time is
NR then the estimated time flag
must be FALSE and the time must be NULL.[132][133] It is required that there be a record of whether
the start and stop times are estimated when there are start
and stop times -- the Start and
Stop columns cannot be non-NULL
when the Btimeest and Etimeest columns, respectively, are
NULL.[134] It is required that there be a record of the
source of the start and stop times when there are start and
stop times -- the Bsource and
Esource values must be NULL
unless, respectively, the Btimeest
and Etimeest values are
non-NULL.
SWERB_BES rows are automatically sequenced when no Seq is specified[135]by Start value,
unless the Start value is NULL
in which case they are sequenced last of all existing
SWERB_BES rows for the group/day when initially inserted and
otherwise not automatically sequenced.[136] In the case of a tie the automatic sequencing
places the newly inserted row[137] last among the rows that are tied. Seq values may be manually assigned so
long as the manual sequencing does not result in
out-of-order Start values, or in
those cases where Start is
NULL, so long as the manually assigned sequence number is
less than or equal to that which would be automatically
assigned.[138]
As expected, changing the Start value (via a SWERB_DATA row with an Event value which indicates the start of observation) will automatically change the Seq value. Should there be other SWERB_BES rows for that group/day with the same SWERB_BEs-Start value the newly changed row will be be sequenced after the existing rows.[139]
Every bout of observation must have exactly one
beginning -- there must be exactly one related row on
SWERB_DATA with an Event of
B. Every bout of
observation must have exactly one end -- there must be exactly
one related row on SWERB_DATA with an Event of
E. These requirements are
enforced on transaction commit, so the
SWERB_BE row and the begin and end SWERB_DATA rows must all be created within a
single transaction. The system will
generate a warning when there are no observations in a bout of
observation -- when there are no related SWERB_DATA rows with Event values other than
B and
E.
The focal group, Focal_grp, must be in existence, based on GROUPS.Start and GROUPS.Cease_To_Exist, on the date of the observation.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and may be used to refer to a particular bout of uninterrupted observation.
This column is automatically maintained by the
database[140], cannot be changed, and must not be
NULL.
The id of the SWERB_DEPARTS_DATA row
representing the departure from camp of the observation
team. This column cannot be changed.[141] This column must not be NULL.
The group under observation. The legal values for
this column are from the Gid column
of the GROUPS table. This column cannot
be changed.[142]This column may not be NULL.
The time the bout of observation started. The time
may not be before 05:00 and may
not be after 20:00. The time must
be on the minute mark; the seconds must be zero. This
column may be NULL when the start of observation is
unknown.
TRUE when the Start
value is an estimation of the time the daily observation of
the group began. FALSE otherwise. This column should be
NULL when the Start time is
the start of a uninterrupted bout of observation but is not
the start of the day's observation of a group.
The source of the data used to estimate the Start value when that value is
estimated and represents the start of the day's observation
of the group -- how the start of the daily observation of
the group was estimated. The legal values of this column
are defined by the SWERB_TIME_SOURCES
table. This column must be NULL when the Start time is the start of a
uninterrupted bout of observation but is not the start of
the day's observation of a group.
The time the bout of observation ended. The time may
not be before 05:00 and may not
be after 20:00. The time must be
on the minute mark; the seconds must be zero. This column
may be NULL when the end of observation is unknown.
TRUE when the Stop value
is an estimation of the time the daily observation of the
group began. FALSE otherwise. This column should be
NULL when the Stop time is the
end of a uninterrupted bout of observation but is not the
end of the day's observation of a group.
The source of the data used to estimate the Stop value when that value is
estimated and represents the end of the day's observation of
the group -- how the end of the daily observation of the
group was estimated. The legal values of this column are
defined by the SWERB_TIME_SOURCES table.
This column must be NULL when the Stop time is the end of a
uninterrupted bout of observation but is not the end of the
day's observation of a group.
A sequence number indicating the ordering of the bouts
of uninterrupted observation of each group each day. The first
bout of observation for the group for the day has a Seq
value of 1, the second a value of
2, etc.
The system automatically re-computes Seq values to
ensure that they are contiguous and begin with
1. See the overview of the SWERB_BES table and the Automatic Sequencing section for further
information.
A boolean value. TRUE means that the bout of
observation counts toward total observer effort. FALSE
means that the bout is concurrent with another bout of
observation by the same team and should not count toward
observer effort.
This column cannot be NULL.
Notes, if any, on the bout of observation. This column may not be empty, it must contain characters, and it must contain at least one non-whitespace character.
This column may be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every event related to group-level geolocation.[143]Such events geolocate a group upon the occurrence of a significant activity, including but not limited to ascent, descent, and drinking. Other events include geolocation at regular intervals and the begin and end of each bout of uninterrupted observation.
Note
The typical Babase user may find the SWERB view to be easier to query than SWERB_DATA and its related tables. It may be easier to use the SWERB_DATA_XY view to maintain SWERB_DATA than it is to modify the table content directly.
Rows with an Event value of
O or
P are not part of an
observation bout of the focal group and so, unless the
observed group is a Subgroup[144] or is the unknown group[145], must have a Seen_grp
value which differs from that of the group under observation
-- the SWERB_BES.Focal_grp value of the related SWERB_BES row. Likewise, rows which do not have
an Event value of
O or
P must have a Seen_grp value of the group under
observation -- a value which equals the SWERB_BES.Focal_grp
value in the related SWERB_BES
row.[146]The system will generate a warning when the SWERB_DATA row is for a non-focal group and the
observed group is a subgroup and the observed group is the
same as the focal group -- when Event is
O and Subgroup is TRUE and SWERB_DATA.Seen_grp
is the same as the related SWERB_BES.Focal_grp.
Per bout of observation, per BEId, there must be exactly one
SWERB_DATA row recording the start and exactly one recording
the finish of the bout -- exactly one SWERB_DATA row having an
Event value of
B and exactly one having a
E value, respectively.
The time of the observation must be between the start
and stop times of the bout of observation -- the Time value must be between (inclusive)
the related SWERB_BES.Start and SWERB_BES.Stop values.
Because SWERB_BES.Start may be NULL the Time value is also checked to be sure
that it's not before the time the observation team departed
from camp, before SWERB_DEPARTS_DATA.Time. Because SWERB_DEPARTS_DATA.Time may also be NULL the
Time value is checked to be sure
that it is not before 05:00. Because SWERB_BES.Stop may be
NULL the Time value is checked
to be sure that it is not after 20:00.
The date portion of the GPS_Datetime value must be the date of the observation team's departure from camp -- must equal the related SWERB_DEPARTS_DATA.Date value. The waypoint time recorded by the operator cannot be more than 15 minutes before the actual time the observation was taken -- the Time value cannot be more than 15 minutes before the time portion of the GPS_Datetime value. The exception to this rule is when a group drinks from a water hole; for these water hole events, the waypoint time cannot be more than 30 minutes minutes before the actual time the observation was taken. The waypoint time recorded by the operator cannot be more than 5 minutes after the actual time the observation was taken -- the Time value cannot be more than 5 minutes after the time portion of the GPS_Datetime value.
The Quad column records
group location based on map quadrants and is used only in
older data. Data recorded after 1994-09-30, rows
associated with SWERB_DEPARTS_DATA rows with
Date values after
1994-09-30, must have NULL Quad values. GPS units were used in
later SWERB data collection so data recorded before
1993-09-01, rows associated with SWERB_DEPARTS_DATA rows having Date values before
1993-09-01, must have NULL XYLoc values.
Only data collected using GPS units have altitude, PDOP,
accuracy, a GPS timestamp, or Garmincode values -- when the
XYLoc column is NULL then the
Altitude, PDOP, Accuracy GPS_Datetime, and Garmincode values must also be
NULL.
The observed lone animal must be NULL unless the
waypoint is an observation of a lone animal/non-focal group
— Lone_Animal must be
NULL unless Event is
O.
Note
An other group observation of an unknown lone animal
is recorded in a SWERB_DATA row having a NULL Lone_Animal value and a Seen_grp value of
10.0 (the group denoting a lone
animal).
The observed predator must be NULL unless the waypoint
is an observation of a predator — the Predator must be NULL unless the
Event is
P, in which case the Predator must not be NULL.
The observer's distance from the observed lone animal,
predator, or non-focal group must be NULL unless the
waypoint is an observation of a lone animal, predator, or
non-focal group -- Ogdistance
must be NULL unless Event is
O or
P.
Note
Through the end of 2022, the observers' protocol for
recording this distance was either been poorly defined or
poorly adhered-to. (It is unclear which.) Distances were
occasionally recorded but usually not. It is unclear what
decisions were made at the time that might decide whether or
not to record this distance. To avoid fallacious assumptions
about the nature of the data, all distances recorded before
01 Jan 2023 have been manually set to NULL.
In case someone wants to use the SWERB_DATA_HISTORY
table to retrieve the once-present distances, they were set
to NULL at 2023-03-22
00:20:44.206126+03 (Nairobi time).
The observed group, Seen_grp, must be in existence, based on GROUPS.Start and GROUPS.Cease_To_Exist, on the date of the observation.
An observed lone animal, Lone_Animal, must have already entered the study population and must be alive on the date of observation -- the SWERB_DEPARTS_DATA.Date related to the SWERB_DATA row must be between individual's related Entrydate and Statdate, inclusive. The system will return a warning if the related Date is before the individual's LatestBirth.
The system will generate a warning if a lone animal is a male and is observed more than 60 days before his assigned dispersal date -- before DISPERSEDATES.Dispersed.
When a lone individual is observed, the observed group
must be the group reserved for lone animals -- when
SWERB_DATA.Lone_Animal is
non-NULL then SWERB_DATA.Seen_grp must be
10.0.
Caution
Interpolation does not reference SWERB data when making its computations. Consequently the MEMBERS table does not reflect SWERB sightings of lone individuals -- unless those sightings are otherwise recorded in the DEMOG table.
When a predator is observed, the observed group must be
the group reserved for predator sightings -- when
SWERB_DATA.Predator is non-NULL
then SWERB_DATA.Seen_grp must be
99.0.
Note
It is not possible from these data to determine the number (quantity) of predators observed. Information like this is recorded, but not in the GPS units[147]. See the Amboseli Baboon Research Project Monitoring Guide for more information.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and may be used to refer to a particular GPS event.
This column is automatically maintained by the
database, cannot be changed, and must not be NULL.
The id of the SWERB_BES row
representing the bout of uninterrupted observation of which
the SWERB_DATA row is a part. This column cannot be changed
and must not be NULL.
The group under observation. Note that this is not always the focal group which the observation team set out to observe. For further details see the Protocol for Data Management: Amboseli Baboon Project. The legal values for this column are from the Gid column of the GROUPS table.
This column may not be NULL.
The BIOGRAPH.Sname of the observed lone animal.
This column may be NULL to indicate either that a
lone animal was not observed or that an unknown lone animal
was observed.
A code indicating what sort of event the row represents. The following codes are defined:
BThe row represents the beginning of a bout of uninterrupted observation of the focal group.
EThe row represents the end of a bout of uninterrupted observation of the focal group.
HThe row represents an observation of the focal group. These occur on half hourly or hourly intervals, depending on the protocol used to record the data.
WThe row records the focal group's drinking.
OThe row represents the observation of a non-focal group or lone animal.
PThe row represents a sighting of a predator.
This column may not be NULL.
The time of the observation. This is usually the time
manually entered by the observer but in those cases where
the observer does not enter a time (such as begin and end
rows) the SWERB_UPLOAD view may use GPS
supplied information to calculate a time. See the section
on the SWERB_UPLOAD.Description column. The time must
be on the minute mark; the seconds must be zero. This
column may be NULL when the time is not known.
The map quadrant of the seen group's location, when recorded in the field. The legal values for this column are from the Quad column of the QUAD_DATA table.
This column may be NULL.
XYLoc (X and Y WGS 1984 UTM Zone 37South coordinates)
The X and Y WGS
1984 UTM Zone 37South coordinates of the seen group. This
column may be NULL.
See the SWERB Data overview for more information.
The altitude, in meters, of the landscape on which the
seen group is
located. This column may be NULL.
See the SWERB Data overview for more information.
The amount of error reported as positional dilution of
precision. This column may be NULL when there is no PDOP
information.
See the SWERB Data overview for more information.
The accuracy of the GPS reading, in meters. This
column may be NULL when there is no accuracy information
in meters.
See the SWERB Data overview for more information.
TRUE when the observation is of a subgroup, FALSE
when not.
Note that the field team cannot always record subgroup information and the value in this column is therefore sometimes determined heuristically[148] when the data is uploaded by the SWERB_UPLOAD view.
This column must not be NULL.
The distance, in meters, between the observer and the
observed non-focal group or the observer and the observed
lone animal. This value must be a 3
digit non-negative integer that is also a mulitple of
0.
This column may be NULL when the observers did not
record an Ogdistance (i.e. NULL values are not to be
confused with zero distance).
The date and time automatically supplied by the GPS
unit at the time the waypoint was recorded. For further
information on when this column is NULL and when
non-NULL see the SWERB Data
overview.
This column may be NULL.
The information manually entered by the observer into
the GPS unit as a coded waypoint that describe the SWERB
data being recorded. This column may be empty, it need not contain characters,
but it may not contain only whitespace characters. For further
information on when this column is NULL and when
non-NULL see the SWERB Data
overview.
This column may be NULL. See the SWERB Data
overview for more information.
The PREDATORS.Predator code of the observed predator.
This column may be NULL, when this row is not for a
predator sighting.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every departure from camp of every observation team, for those observation teams which have collected SWERB data.
The Time value may
not be NULL when there is a related SWERB_DEPARTS_GPS row -- data collected using the
GPS units must have a non-NULL time.
One observer may not depart camp on the same day at the
same time with two different observation teams -- the
combination of SWERB_DEPARTS_DATA.Date, SWERB_DEPARTS_DATA.Time, and SWERB_OBSERVERS.Observer, when all are non-NULL,
must be unique.
The system will generate a warning for SWERB_DEPARTS_DATA rows having a Date after 1994-09-30 that do not also have a related SWERB_DEPARTS_GPS row.
The system will generate a warning for SWERB_DEPARTS_DATA rows for which no SWERB data was collected; that do not have a related SWERB_BES row.
Note
The SWERB_DEPARTS view can be used to maintain the SWERB_DEPARTS_DATA table. This view may also be more useful than the table when querying.
Warning
At the time of this writing departure data prior to about March of 2011 is not in the database. The process involved in loading historical data fabricates (departure date excepted, the actual departure date is used) the minimal required departure information. The early process used by the Data Manager involving loading data from the GPS units sometimes involved removing departure information. For further information and exact dates see the Data Manager's [Process for Uploading SWERB] document.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and may be used to refer to a particular departure from camp of a particular observation team.
This column is automatically maintained by the
database, cannot be changed, and must not be NULL.
The time of departure. The time may not be before
04:00 and may not be after
20:00. The system will generate a
warning if the time is before
05:00 or after
14:30. The time must
be on the minute mark; the seconds must be zero. This
column may be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every departure from
camp of every observation team, for those observation teams
which have collected SWERB data using GPS units. This table
is an extension of the SWERB_DEPARTS_DATA
that contains the additional information collected when a GPS
unit is used to record the departure. There is at most one
row in this table for every row in SWERB_DEPARTS_DATA. When a row exists it contains
the information involving the GPS unit used by the observation
team on that day. All SWERB_DEPARTS_DATA
rows having associated SWERB_DEPARTS_GPS rows must have SWERB_DEPARTS_DATA.Date values on or after
1993-09-01.
The date of departure (SWERB_DEPARTS_DATA.Date) must be between the SWERB_DEPARTS_GPS' Start and Finish dates, inclusive.
Note
The SWERB_DEPARTS view can be used to maintain the SWERB_DEPARTS_GPS table. This view may also be more useful than the table when querying.
The system will generate a warning when there is more than one departure per GPS unit per day.
The id of the SWERB_DEPARTS_DATA row
representing the departure from camp of the observation
team. This column cannot be changed and must not be
NULL.
XYLoc (X and Y WGS 1984 UTM Zone 37South coordinates)
The X and Y WGS
1984 UTM Zone 37South coordinates at departure. This
column must not be NULL.
See the SWERB Data overview for more information.
The altitude in meters of the GPS unit. This column
may be NULL.
See the SWERB Data overview for more information.
The error reported as positional dilution of
precision. This column may be NULL.
See the SWERB Data overview for more information.
The error reported in meters. This column may be
NULL.
See the SWERB Data overview for more information.
The identifier of the GPS device (the GPS_UNITS.GPS) used by the observation team. The legal values of this column are defined by the GPS_UNITS support table.
This column must not be NULL.
The information manually entered into the waypoint by
the observer. This is a set of, mostly, single character
codes that describe the SWERB data being recorded.
This column may be empty, it need not contain characters,
but it may not contain only whitespace characters. For further information on when this column
is NULL and when non-NULL see the SWERB Data overview.
This column may be NULL. See the SWERB Data
overview for more information.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every geolocated physical object, that is, for every grove and waterhole.[149]
Caution
This table may contain one row with special meaning.
The SWERB_GWS row with a Loc
value of UNK represents
the unknown grove -- a grove with special properties. When a
SWERB_GWS row exists with a SWERB_GWs-Loc value of
UNK then the Type value must be
G (grove). No trees
may be located in the unknown grove -- TREES.Loc may not be
UNK. The unknown grove
may not be located anywhere -- SWERB_GW_LOC_DATA.Loc may not be
UNK. And when it is not
known where a group slept there can be no uncertainty
regarding the sleeping grove -- when SWERB_LOC_DATA.Loc is
UNK then SWERB_LOC_DATA.Loc_Status
must be C
(certain).
SWERB_GWS rows that represent groves, those with a
SWERB_GWs-Type of G,
have restrictions on the allowed Loc values due to the data structure
supplied the SWERB_UPLOAD view (the Name column sometimes contains a
grove code prefaced with the letter
“P”). There cannot be two
codes for groves, one which begins with the letter
“P” and another which
consists entirely of the same characters as the first but with
the initial “P”
omitted.[150] Because of this restriction the Babase
administrator is the only user allowed to create Loc values which begin with the letter
“P”.
With the exception of the unknown grove, the system will report a warning when the grove or waterhole has not been geolocated -- when there is no related SWERB_GW_LOC_DATA row.
A unique identifier. Up to 4 alphanumeric non-lowercase characters that uniquely identifies the row and may be used to refer to the grove or waterhole.
This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character. This column cannot be changed and must not
be NULL.
The type of place; whether grove, waterhole, or some other landmark. The legal values for this column are from the Place column of the PLACE_TYPES (codes for various landscape features) table.
This column must not be NULL.
Up to 20 characters of alternative name for the grove or waterhole.
This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character. This column may be NULL.
The date when the grove or waterhole was named. This
date cannot be before
1981-11-01.
This column must not be NULL.
The date of last known use after which the resource became permanently unavailable.
This column may be NULL when observations are
ongoing or the row represents an object that cannot become
unavailable.
Textual notes on the grove or waterhole, if any.
This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character. This column may be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for each time a location of a place, a grove or waterhole is recorded. Any given grove or waterhole may have its location recorded more than once.
Note
The typical Babase user may find the SWERB_GW_LOCS view to be easier to query than SWERB_GW_LOC_DATA and its related tables. It may be easier to use the SWERB_GW_LOC_DATA_XY view to maintain SWERB_GW_LOC_DATA than it is to modify the table content directly.
The date related to the location (SWERB_GW_LOC_DATA.Date) may not be before the grove or waterhole was first observed, may not be before the related SWERB_GWS.Start value. The date related to the location (SWERB_GW_LOC_DATA.Date) may not be after the grove or waterhole ceases existance, may not be after the related SWERB_GWS.Finish value.
The Quad column
records group location based on map quadrants and is used only
in older data. Data recorded after 1994-09-30, rows with
Date values after
1994-09-30, must have NULL Quad values. GPS units were used in
later SWERB data collection so data recorded before
1993-09-01, rows having Date values before
1993-09-01, must have NULL XYLoc values, unless the UTM XY
coordinates were obtained through other means (XYSource is non-NULL).
There can only be a source for the recorded X and Y
coordinates when there are recorded UTM coordinates -- the
XYSource value may be
non-NULL only when XYLoc
is non-NULL. There must be X and Y UTM coordinates when
there is a recorded source for the X and Y coodinates -- XYLoc must be non-NULL when
XYSource is
non-NULL.
Only data collected using GPS units have altitude, PDOP,
accuracy, and GPS values -- when the XYLoc column is NULL then the
Altitude, PDOP, Accuracy, GPS values must also be
NULL.
The GPS unit used to make the observation must be in service on the date of the observation -- the date of the observation (Date) must be between the SWERB_DEPARTS_GPS' Start and Finish dates, inclusive, of the related GPS_UNITS row.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and may be used to refer to an observation which recorded the location of a particular grove or waterhole.
This column is automatically maintained by the
database and must not be NULL.
The SWERB_GWS.Loc of the grove or waterhole associated with the recorded location.
This column must not be NULL.
The date related to the location. This is either the
date the location was caculated or an observation date. See
the Protocol for Data
Management: Amboseli Baboon Project for further information. This column must
not be NULL.
The time of the observation. When the data are taken
with a GPS unit this is the time recorded by the GPS unit.
The time cannot be before 05:00
and cannot be after 20:00. The
time must be on the minute mark; the seconds must be zero.
This column may be NULL when the time is not known.
The map quadrant of the grove or waterhole's location, when recorded. The legal values for this column are from the Quad column of the QUAD_DATA table.
This column may be NULL.
The source of the UTM coodinate data. The legal values for this column are from the XYSource column of the SWERB_XYSOURCES (SWERB Time Sources) table.
This column may be NULL.
XYLoc (X and Y WGS 1984 UTM Zone 37South coordinates)
The X and Y WGS
1984 UTM Zone 37South coordinates of the grove or
waterhole. This column may be NULL.
See the SWERB Data overview for more information.
The altitude, in meters, of the grove or waterhole.
This column may be NULL.
See the SWERB Data overview for more information.
The error reported as positional dilution of
precision. This column may be NULL when there is no PDOP
information.
See the SWERB Data overview for more information.
The error reported in meters. This column may be
NULL when there is no accuracy information in
meters.
See the SWERB Data overview for more information.
The identifier of the GPS device (the GPS_UNITS.GPS) used in the observation. The legal values of this column are defined by the GPS_UNITS support table.
This column may be NULL.
See the SWERB Data overview for more information.
Textual notes regarding the record of the grove or waterhole's location, if any.
This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character. This column may be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row every time a group is observed at a geolocated physical object, i.e. at a grove or a waterhole or, possibly, some other physical landmark.[151]
SWERB_LOC_DATA rows must place a group at a single location -- each SWERB_DATA row has at most one related SWERB_LOC_DATA row. In effect, SWERB_LOC_DATA extends SWERB_DATA with additional columns.
Descent from, or ascent into, groves is indicated in the
ADcode column. To indicate a
descent, the ADCode value must relate to an ADCODES row a ADN value
of D. To indicate an ascent, the ADCode
value must relate to an ADCODES row a ADN value of A.
The observations recording descent from or ascent into
sleeping groves must be related to groves (the related SWERB_GWS rows must have a Type of
G).
The SWERB_DATA row representing the
"begin" of the team's first bout of observation (the bout with
the smallest SWERB_BES.Seq value) of any group (except the
unknown group, group
9.0)[152] in a day must be related to a SWERB_LOC_DATA row
recording descent from a sleeping grove. This enforces the
requirement that a day's observations of a group must include
the group's descent from exactly one
grove (possibly the unknown grove). A group can be recorded
as descending from more than one grove, but only when all of
the descents are by subgroups (the related SWERB_DATA.Subgroup
is TRUE), or all but one of the descents are by subgroups
and those subgroup descents are from the
unknown grove.
Similarly, the SWERB_DATA row
representing the "end" of the team's final bout of observation
(the bout with the greatest SWERB_BES.Seq value) of any group (except the
unknown group, group
9.0)[153] in a day must be related to a SWERB_LOC_DATA row
recording ascent into a sleeping grove. This enforces the
requirement that a day's observations of a group must include
the group's ascent into exactly one grove
(possibly the unknown grove). A group can be recorded as
ascending into more than one grove, but only when all of the
ascents are by subgroups (the related SWERB_DATA.Subgroup
is TRUE), or all but one of the ascents are by subgroups and
those subgroup ascents are into the unknown grove. The
database rules that enforce these "ascent into sleeping grove"
rules are checked at transaction
commit.[154]
Tip
When a group splits into subgroups and descends from or ascends into multiple groves there must be a separate bout of observation, another SWERB_BES row, to record the location of each subgroup.
Whether a SWERB_LOC_DATA row must have a NULL ADtime value or must have a
non-NULL ADtime value is
determined by the related ADCODES.Time flag.[155] Ascent and descent times related to a bout of
observation cannot be before the beginning of the bout of
observation -- SWERB_LOC_DATA.ADtime cannot be before the related
SWERB_BES.Start
time.[156] The database rules that enforce ADtime values are checked at transaction commit.[157]
Note
Descent and ascent times are recorded manually; they are not taken from the timestamps supplied by the GPS units. This necessitates additional columns for descent and ascent information. For further information see the Amboseli Baboon Research Project Monitoring Guide.
Caution
An ascent or descent might occur during a bout of
observation without being recognized or recorded; an ascent
recorded as "after departure" or a descent "before arrival"
might have actually occurred while the observers were
present. Users can safely assume that any ascents/descents
with a known (non-NULL) ADtime are accurate, but users
should not assume that a morning descent indicated as before
arrival (ADcode =
BA) really did occur before the bout's
start time, nor that an evening ascent indicated as after
departure (ADcode =
AD) really did occur after the bout's
stop time.
When the location is the unknown grove, status of that
location must be 'certain'. That is, when the Loc value is
UNK then the Loc_Status value must be
C.
Babase allows SWERB data to record group presence at
arbitrary landmarks, but some possibilities are rare and
result in a warning. The system will issue a warning when a
group is located at a waterhole but the recorded activity is
not “water” (when the SWERB_GWS
row's Type is
W but the related SWERB_DATA row's Event value is not
W).
SWERB_DATA rows representing
observation of a group drinking at a waterhole must be related
to waterholes. That is, when SWERB_DATA.Event is
W there must be a related
SWERB_GWS row, even if it is the generic and
non-specific row which represents all rainpools, and the
related SWERB_GWS row must have a Type value of
W. In some cases this
check is at transaction commit time and in
other cases not.
Rows that record a drinking event -- those related to
SWERB_DATA rows which have
W Event values -- must have
SWERB_LOC_DATA.ADcode values
that indicate no involvement with a sleeping grove; the
related ADCODES row must have a ADN value of
N.
Groups may not be located at a place before observations began at the place or after observations ended at the place. That is, the SWERB_DEPARTS_DATA.Date related to the SWERB_DATA row referenced by the SWERB_LOC_DATA.SWId value must not be before the related SWERB_GWS.Start value and must not be after the related SWERB_GWS.Finish value.
The number that uniquely identifies the row and may be used to refer to an observation of a group at a particular time at a particular grove or waterhole. This is also the SWERB_DATA.SWId identifying the group, place, and time of the observation.
This column must not be NULL and cannot be
changed.
The SWERB_GWS.Loc of the object (grove, waterhole, or landmark) where the group was observed.
This column must not be NULL.
A code representing the nature of the relationship between the baboon group and the landscape feature at which the SWERB_LOC_DATA row places the group. The legal values of this column are defined by the ADCODES support table.[158]
This column must not be NULL.
The SWERB_LOC_STATUSES.Loc_Status value indicating
the status of this observation of the location on record
(this row's Loc). Usually, this will indicate whether the
observers actually saw the group at the location or inferred
that the group was there. For instance, if the group is
still in a sleeping grove when the observers arrive then
they will be "certain" about that grove (Loc_Status =
C), but if the group is
walking away from the grove when the observers arrive then
they may indicate the grove as 'probable'(Loc_Status =
P).
Note
Although the database supports degrees of certainty with respect to any group location, in practical terms the only time that there will be any degree of uncertainty will involve sleeping groves. This is for two reasons. First, at present the only provision in the Amboseli Baboon Research Project Monitoring Guide involving uncertainty is with respect to sleeping groves. Second, the SWERB_UPLOAD will only ever enter an indication of uncertainty into the database when the location is a sleeping grove.[159]
This column may not be NULL.
The median time of group decent from or ascent into a
sleeping grove. See the Amboseli
Baboon Research Project Monitoring Guide for information
regarding how median descent and ascent times are
determined. The time may not be before
05:00 and may not be after
20:00. The time must be on the
minute mark; the seconds must be zero. This column may be
NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
The SWERB data collection protocol sometimes requires 2 GPS waypoint entries to record a group's presence at a physical landscape feature. (At the time of this writing descent from and ascent into sleeping groves requires 2 GPS waypoint entries.) This table contains one row every time a group is observed at a geolocated landscape feature and 2 GPS waypoints are required to record the data. The rows of this table contain the information stored in the second GPS waypoint, information automatically generated by the GPS unit or manually entered into the GPS unit, that otherwise have no place in the database.
Note
It may be easier to use the SWERB_LOC_GPS_XY view to maintain SWERB_LOC_GPS table than it is to modify the table content directly.
The SWERB_LOC_GPS table extends the SWERB_LOC_DATA table[160] with additional columns; SWERB_LOC_GPS contains at most one row for every row in SWERB_LOC_DATA.
As described in the SWERB Data overview above, data was first obtained directly from the GPS units on 2004-01-01. Consequently, this table cannot contain rows dated earlier than 2004-01-01.
The number that uniquely identifies the row and may be used to refer to the GPS information involving an observation of a group at a particular time at a particular grove or waterhole. This is also the SWERB_DATA.SWId value, identifying the group, place, and time of the observation, and the SWERB_LOC_DATA.SWId value, identifying the placement of the group at a landscape feature.
This column must not be NULL and cannot be
changed.
XYLoc (X and Y WGS 1984 UTM Zone 37South coordinates)
The X and Y WGS
1984 UTM Zone 37South coordinates of the SWERB_DATA.seen group. This
column may not be NULL.
See the SWERB Data overview for more information.
The altitude, in meters, of the landscape on which the
seen group is
located. This column may be NULL.
See the SWERB Data overview for more information.
The amount of error reported as positional dilution of
precision. This column may be NULL when there is no PDOP
information.
See the SWERB Data overview for more information.
The accuracy of the GPS reading, in meters. This
column may be NULL when there is no accuracy information
in meters.
See the SWERB Data overview for more information.
The date and time automatically supplied by the GPS
unit at the time the waypoint was recorded. This column may
not be NULL.
This column may be NULL.
The information manually entered by the observer into
the GPS unit as a coded waypoint that describe the SWERB
data being recorded. This column may be empty, it need not contain characters,
but it may not contain only whitespace characters. This column may not be
NULL, although it may be a string 0 characters long.
See the SWERB Data
overview for more information.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
For teams collecting SWERB data this table contains one row for every departure from camp of every member of the departing observation team for those team members who drive or record data.
The system will generate a warning for those SWERB_DEPARTS_DATA rows without at least one related row in SWERB_OBSERVERS.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and may be used to refer to a particular observer's departure from camp as part of a particular observation team.
This column is automatically maintained by the
database, cannot be changed, and must not be NULL.
The id of the SWERB_DEPARTS_DATA row
representing the departure from camp of the observer's
observation team. This column must not be NULL.
Initials of the observer. The legal values of this column are defined by the OBSERVERS support table.
This column must not be NULL.
The role assumed by the member of the SWERB observation team. The legal values of this column are defined by the OBSERVER_ROLES support table.
This column must not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every tree in the tree monitoring project.
Trees can only be located in groves -- the value of the
TREES.Loc column must reference a
SWERB_GWS row which has a SWERB_GWS.Type of
G (Grove).
Tree numbers are unique within each grove. The combination of Loc and Tree must be unique.
A unique identifier. This is an automatically generated sequential number that uniquely identifies the row and may be used to refer to a particular tree.
This column is automatically maintained by the
database, cannot be changed, and must not be NULL.
The identifier of the grove, a SWERB_GWS.Loc value, in which the tree is located.
This column must not be NULL.
The integer used to uniquely identify a tree within a particular grove.
This column must not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
The data in this section are collected from manually read instruments, with one notable exception: the DIGITAL_WEATHER table contains data from electronic instruments that record weather data automatically.
Tip
The MIN_MAXS view provides a way to view all the tables containing manually collected weather data at once, with each weather data collection event appearing as a single row.
Tip
You should never assume that the data in these tables are a highly precise and accurate record of the rainfall and temperature experienced by the baboons in Amboseli. Care is taken to clean and curate the data as much as possible. However, undetected errors associated with the observers or the instruments may occur from time to time. Furthermore, when instruments are changed (for instance, one min-max thermometer is substituted for another) or when the location of the weather station changes, this can affect the resulting records. In general, relying on a single weather station is not a best practice in ecological analysis. Therefore, we recommend that you consider also examining the data from other weather measurement systems (e.g. MERRA-2 or Berkeley Earth), especially if your analyses find something remarkable in these data.
Note
The weather-related tables contain weather-related information and so do not directly relate to any of the baboon information contained in Babase.
Equipment irregularities or
failures occasionally occur, resulting in periods of data whose
accuracy is uncertain. Data from these periods that are not
known to be incorrect may still be retained
for reference[161], but their veracity is uncertain. Accordingly, some
of the tables discussed in this section include columns that
indicate whether a temperature reading has been "curated". When
TRUE, the reading is considered reliable for general use and
therefore will appear in the CURATED_TEMPS
view. When FALSE, the reliability of the reading is in
question and it should generally be excluded from analytical use
unless there is a specific reason to include it.
Tip
To use only temperature data that have been "curated", use the CURATED_TEMPS view.
This table contains one row for every time a rain gauge reading is recorded. There can be at most one RAINGAUGES row per WREADINGS row.
The identifier of the meteorological collection event during which the rain gauge was read. Must be a value contained in the WRid column of a row on the WREADINGS table, and the associated row may not be associated with any other row in RAINGAUGES.
This column cannot be changed; and must not be
NULL.
The interval, in an integral number of seconds, since the previous rain gauge collection event.
This column is automatically maintained by the
database and cannot be changed. This column must not be
NULL.
Caution
When the WREADINGS.WRdaytime values used to compute RGspan are not integral, the resulting RGspan value is rounded to the nearest second. Values of .5 seconds are rounded to the nearest even number of seconds.
Warning
When a new row is inserted the value of this column
is silently ignored and an automatically computed value is
used in its place. It is best to omit this column from
the inserted data (or specify the NULL value).
Whether or not any estimated WREADINGS.WRdaytime
values were used in the computation of the RGspan column. TRUE if any of the
relevant WREADINGS.Estdaytime values are true, FALSE
otherwise.
This column is automatically maintained by the
database and cannot be changed. This column must not be
NULL.
Warning
When a new row is inserted the value of this column
is silently ignored and an automatically computed value is
used in its place. It is best to omit this column from
the inserted data (or specify the NULL value).
The measurement of rain accumulated since the last time the rain gauge was read. In millimeters stored using a data type having a precision of 0.1 millimeter. For the precision and accuracy of the data itself see the Amboseli Baboon Research Project Monitoring Guide.
This column must be non-negative and may not be more
than 200.0. This column may not
be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every time a rain gauge is installed. There can be no RAINGAUGES rows recording rain gauge measurements at any given weather station (WSTATIONS) unless there is a prior record of a rain gauge installation in RGSETUPS.
Rain gauge measurements are only meaningful when it is known how long the rain has been collected. In the event that, e.g., an elephant steps on the rainguage, there will be a period of time until the rain gauge is replaced. The first reading of the replacement rain gauge is not a measurement of rain since the last rainguage reading, but is instead a measurement of the rain collected since the replacement rain gauge was installed. The RGSETUPS table allows the system to compute RAINGAUGES.RGspan intervals when rain gauges are replaced, first installed, or after an interval of corrupted measurements.[162]
There cannot be a RGSETUPS row and a RAINGAUGES row for the same location at the same time.
The combination of RGSdaytime and Wstation must be unique.
A unique positive integer representing the rain gauge setup event.
This column is automatically maintained by the
database, cannot be changed, and must not be NULL.
Code indicating the station at which the rain gauge was installed. Must be a value on the WSTATIONS table.
This column cannot be changed and must not be
NULL.
TRUE when the RGSdaytime
column contains an estimated time. FALSE when the RGSdaytime column is an accurate record
of the time the rain gauge was installed.
Initials of the person who collected the data. Must be a value contained in the Initials column of a row on the OBSERVERS table.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every time a minimum temperature reading was recorded. There can be at most one TEMPMINS row for every WREADINGS row.
The Tempmin column has one
decimal point of precision, but its accuracy is lower.
Readings are only considered reliable to the nearest 0.5
degrees, so the digit to the right of the decimal point must
be either a 0 or a
5.
The Curated column is used to indicate the reliability of this minimum temperature reading, as discussed above. Temperature readings collected manually tend to be more vulnerable to human error, so the default value for Curated is FALSE.
It is unlikely but possible that a minimum temperature
is curated, but the maximum temperature from the same reading
is not. The system will return a warning for each WRid with a TRUE TEMPMINS.Curated and a
FALSE TEMPMAXS.Curated.
The identifier of the meteorological collection event during which the minimum temperature was read. Must be a value contained in the WRid column of a row on the WREADINGS table, and the associated row may not be associated with any other row in TEMPMINS.
This column cannot be changed; and must not be
NULL.
The minimum temperature recorded since the last minimum temperature reading.
This table must contain a value between
-5 and
35, inclusive of endpoints,
and must not be NULL.
A boolean, indicating whether or not this minimum temperature reading has been curated.
This column cannot be NULL and is FALSE by
default.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every time a maximum temperature reading was recorded. There can be at most one TEMPMAXS row for every WREADINGS row.
In extreme circumstances where a temperature reading is
known to be spurious in some way, it may be desirable to
record a correction or adjustment from the original
temperature. When this is done, the adjusted temperature
should be recorded in the Tempmax
column, and the unadjusted temperature in the Unadjusted_Tempmax column. If no
adjustment has been made, the Unadjusted_Tempmax should be
NULL.
Because a non-NULL Unadjusted_Tempmax indicates that an
adjustment has occurred, the Unadjusted_Tempmax cannot be equal to the
Tempmax.
Both temperature columns have one decimal point of
precision. However, the accuracy of the Unadjusted_Tempmax is lower. Readings
are only considered reliable to the nearest 0.5 degrees, so
its decimal digit must be either a 0 or a
5. The Tempmax
is also generally not more accurate than the nearest half of a
degree, but because it may reflect a corrected or adjusted
value its decimal digit can be any number. The system will
return a warning when the Tempmax
is not a multiple of 0.5.
Values in both of the temperature columns in this table
must be between 10 and
50, inclusive.
Historical Note
Weather station BC1 was positioned too close to the kitchen, resulting in spuriously high Tempmax readings. To correct for this, all Tempmax readings from that weather station have been adjusted by -4.2°C (rounded from -4.245). This adjustment was calculated as the residual + fixed effect from a model of Tempmax as a function of day of the year + random intercept of weather station with only BC1 and BC2, BC3, BC4 combined in the dataset (i.e., Tempmax ∼ day of the year + (1 | Wstation)). Day of the year was included in the model to correct for the fact that BC1 had an overrepresentation of January to June dates compared to the other three BC weather stations. BC5 was not used in the calculation because at the time of calculation there was less than one year of weather data from this station. We also calculated adjustment factors in two alternative ways which yielded extremely similar values: (1) taking the difference between the mean Tempmax of BC1 and mean Tempmax of BC2, BC3, BC4 combined (adjustment factor = -4.29°C) and (2) taking a residual + fixed effect from a model of Tempmax as a function of a fixed intercept + random intercept of weather station with only BC1 and BC2, BC3, BC4 combined in the dataset (i.e., Tempmax ∼ 1 + (1 | Wstation); adjustment factor = -4.28°C).
The Curated column is used to indicate the reliability of this maximum temperature reading, as discussed above. Temperature readings collected manually tend to be more vulnerable to human error, so the default value for Curated is FALSE.
It is unlikely but possible that a maximum temperature
is curated, but the minimum temperature from the same reading
is not. The system will return a warning for each WRid with a TRUE TEMPMAXS.Curated and a
FALSE TEMPMINS.Curated.
The WREADINGS.WRid of the meteorological collection event during which this maximum temperature was read.
This column is unique, cannot be changed, and must
not be NULL.
The maximum temperature recorded since the last maximum temperature reading.
This column may not be NULL.
The original, unadjusted maximum temperature, when the value in the Tempmax column has been adjusted in some way.
This column may be NULL, when the Tempmax has not been adjusted.
A boolean, indicating whether or not this maximum temperature reading has been curated.
This column cannot be NULL and is FALSE by
default.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table records the weather data that are automatically collected each hour by an electronic weather collection instrument.
Note
Originally, this table only contained data from WeatherHawk devices and was therefore named WEATHERHAWK. Likewise, the WEATHER_SOFTWARES table was originally named WEATHERHAWK_SOFTWARES. On 28 Nov 2023, these tables were renamed to reflect that they may also contain data related to other devices. Ideally, the WEATHERHAWK_HISTORY and WEATHERHAWK_SOFTWARES_HISTORY tables should remain in the babase_history schema so that changes to those tables will remain accessible. However, when these tables were renamed their history tables were both empty. There were no archived changes in either table that needed to be preserved, so the old history tables were not retained.
A weather station cannot have more than one reading at the same time. That is, the combination of TimeStamp and WStation must be unique.
Instrument accuracy may not, and probably does not, correspond with the recorded degree of precision. These instruments collect their data in engineering units, which are interpreted and converted to standardized units (degrees, kPa, etc.) by PC software when the data are retrieved from the instrument. Different PC software programs may vary in terms of units used, the number of significant figures employed, or other ways that are not immediately apparent. There are even some values that are simply not recorded by some programs or devices.
Despite hardware and software differences, most measurements saved in this table use a single column and a specified unit. Data managers should ensure that data are converted to the appropriate units, if needed. The allowed precision in these columns — usually a single digit to the right of the decimal — is based on a private message from WeatherHawk's technical support[163], who asserted that this is the maximum plausible precision that WeatherHawk devices are capable of measuring. It is presumed that this is also the maximum plausible precision for other (non-WeatherHawk) devices. This might be more or less precise than the value originally reported by the software.
Tip
Use the WEATHER_SOFTWARES table to see what is known about differences in these programs, including precision of measurements, units used, etc.
The WSoftware column is used to indicate which software was used to generate the data in each row, but the system does not treat data any differently based on this value. Users should be aware of the possibility of differences between programs, and decide for themselves how to handle any possible discrepancies.
Information about the voltage of the device's battery is provided in the BatVolt and BatVolt_Min columns. These values are not directly relevant to weather but can be useful if technical support is needed.
Wind speed may be recorded in km/hr as an integer or m/s
with 1 decimal point of precision, depending on the software
used. The precision difference between these two measures is
large enough that they are divided into separate columns.
Each row must indicate the average wind
speed; exactly one (not both) of the WindSpeed_Avg_Km_Hr and WindSpeed_Avg_M_S columns must not
be NULL. Maximum wind speed is not
required, but when recorded it must be in either the WindSpeed_Max_Km_Hr or WindSpeed_Max_M_S column, but not
both.
Each row must only use a single unit for all of its wind
speed values; when WindSpeed_Avg_Km_Hr is NULL,
WindSpeed_Max_Km_Hr must
also be NULL, and when WindSpeed_Avg_M_S is NULL, WindSpeed_Max_M_S must also be
NULL.
The barometric pressure value provided in this table (Barometer) is corrected, accounting for Amboseli's elevation: ~1130 m. To calculate the uncorrected values, ask a meteorologist.
Note
Prior to Babase 5.5.3, this column contained only UNcorrected values. Those values were corrected simply by adding 12.94503[164]to the uncorrected value.
When devices like these record rainfall, they often use a small "tip bucket" that only records rain when the bucket fills (see the device's user's manual for more information) and which theoretically may contribute to small errors in the accuracy of the measurement. For example, the WeatherHawk used a 1-mm tip bucket. If there is less than 1 mm of rainfall over the course of a given hour, the bucket may not fill up at that time and the rain will not be measured until later or may evaporate before the bucket fills. When there is a gap in the hourly measurements (due to changing out sensors, battery malfunctions, etc.), rainfall data during the down period might not be recorded.
Despite the fact that data are recorded every hour, some devices (e.g. WeatherHawk) do not simply report the amount of rainfall measured in that hour. Instead, these devices report the cumulative amount of rainfall measured since the beginning of the year[165]. That value is recorded in the YearlyRain column, for those devices that report it.
The rainfall for each hour is recorded in the TimeStampRain column. For rows
whose YearlyRain column is
not NULL, this value is the result of a simple calculation:
this row's YearlyRain minus
that of the chronologically previous row.
Note
When this table was first created and only contained data from WeatherHawk devices, the value of the TimeStampRain column was automatically calculated when new rows were added. That is, for a given row, the YearlyRain of the most recent row from the same calendar year and the same WStation was subtracted from the given row's YearlyRain, resulting in the amount of rainfall that was measured since the previous TimeStamp.
After the last WeatherHawk device was retired and data
from other devices began to be added, this automatic
calculation stopped being useful. In Babase 5.5.1, this table's
ability to calculate TimeStampRain from YearlyRain was removed, largely
based on the assumption that future devices are unlikely to
use the dubious YearlyRain
measurement. All previously calculated TimeStampRain values were
not removed, so the TimeStampRain in a row with a
non-NULL YearlyRain can
safely be assumed to be a result of that
functionality.
The amount of rain measured in the year cannot be less
than the amount measured in a single timestamp. That is, when
the YearlyRain is not NULL
it cannot be greater than the TimeStampRain.
Warning
Do not assume that TimeStampRain values always describe a single hour's worth of rain. When one or more hours is absent from the data, the TimeStampRain value is the amount of rainfall measured since the previous row in the same year. Also do not assume that these values describe all of the rain that occurred in the intervening hours. If the device was off or malfunctioning at the time, then actual rainfall may have occurred and/or evaporated without being measured.
The AirTemp_Min_Curated and AirTemp_Max_Curated columns are used to indicate the reliability of the AirTemp_Min and AirTemp_Max (respectively), as discussed above. Temperature readings collected automatically tend to be invulnerable to human error, so the default value for both Curated columns is TRUE.
Every temperature min and max reading must be indicated
as curated or not. That is, the AirTemp_Min and AirTemp_Min_Curated must both be
NULL or both not NULL, and the AirTemp_Max and AirTemp_Max_Curated must both be
NULL or both not NULL.
A unique positive integer identifying the device's meteorological data collection that is recorded in this row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
Date and time of the measurement. Measurements must
be taken on the hour. Minutes,
seconds, microseconds etc must be
0.
Warning
As indicated by the name, this value is a time
stamp. It indicates the end of the period described in
each row, not the beginning. This means that the last
hour of a day will have a TimeStamp from the next day,
e.g. the data from 23:00-23:59 on 31 Dec 1999 will have
a TimeStamp of 2000-01-01
00:00.
This column may not be NULL.
The WEATHER_SOFTWARES.WSoftware value indicating which software was used to generate the data.
This column may not be NULL.
The record number for this line, exported in the software. This appears to be a unique ID number used by the device or the software, or both.
This column may be NULL if the software did not
report this value.
The voltage of the battery at the TimeStamp. Values must be
between 10.00 and
14.00, inclusive.
This column may not be NULL.
The minimum voltage of the battery in this hour.
Values must be between 10.00
and 14.00, inclusive.
This column may be NULL if the software did not
report this value.
Average air temperature for this hour, in degrees
Celsius. Values must be between
-10.0 and
50.0, inclusive.
This column may not be NULL.
Average relative humidity for this hour in percent
humidity. Values must be between
0.0 and
100.0, inclusive.
This column may not be NULL.
Average wind speed for this hour, in km/hr. Values
must be between
0 and
30,
inclusive.
This column may be NULL.
Average wind speed for this hour, in m/s. Values
must be between 0.0
and 15.0,
inclusive.
This column may be NULL.
Solar radiation in Watts per square meter. Values
must be between 0.0 and
2000.0, inclusive.
This column may be NULL if the device did not
report this value or if a reported value was subsequently
recognized as erroneous.
Minimum air temperature for this hour, in degrees
Celsius. Values must be between
-10.0 and
50.0, inclusive.
This column may be NULL if the software did not
report this value.
A time stamp indicating the minute in which the AirTemp_Min occurred.
This column may be NULL if the software did not
report this value.
A boolean, indicating whether or not the AirTemp_Min has been curated.
When AirTemp_Min
is not NULL, this column set to TRUE by
default.
Maximum air temperature for this hour, in degrees
Celsius. Values must be between
-10.0 and
50.0, inclusive.
This column may be NULL if the software did not
report this value.
A time stamp indicating the minute in which the AirTemp_Max occurred.
This column may be NULL if the software did not
report this value.
A boolean, indicating whether or not the AirTemp_Max has been curated.
When AirTemp_Max
is not NULL, this column set to TRUE by
default.
A textual value describing or naming the secondary temperature probe, if used.
This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character. This column may be NULL if a secondary
temperature probe was not used. If there are any
secondary temperature values — if any of the AirTemp_Avg_Secondary, AirTemp_Min_Secondary, or
AirTemp_Max_Secondary
columns are not NULL — then this column cannot be
NULL.
Average air temperature for this hour, in degrees
Celsius, measured by the secondary temperature probe.
Values must be between -10.0
and 50.0, inclusive.
This column may be NULL if the software did not
report this value.
Minimum air temperature for this hour, in degrees
Celsius, measured by the secondary temperature probe.
Values must be between -10.0
and 50.0, inclusive.
This column may be NULL if the software did not
report this value.
A time stamp indicating the minute in which the AirTemp_Min_Secondary occurred.
This column may be NULL if the software did not
report this value.
Maximum air temperature for this hour, in degrees
Celsius, measured by the secondary temperature probe.
Values must be between -10.0
and 50.0, inclusive.
This column may be NULL if the software did not
report this value.
A time stamp indicating the minute in which the AirTemp_Max_Secondary occurred.
This column may be NULL if the software did not
report this value.
Wind direction in degrees from North. Values must
be between 0.0 and
360.0, inclusive.
Caution
The values of 0.0
and 360.0 represent the
same direction. There's no telling if one or the other
of them means something special, like “no
measurement”. If they really do represent the
same direction then we should probably change the rules
and adjust the data values so that legal values are
between 0 and
359.
This column may not be NULL.
Maximum wind speed for this hour, in km/hr. Values
must be between
0 and
30,
inclusive.
This column may be NULL if the software did not
report this value.
Maximum wind speed for this hour, in m/s. Values
must be between 0.0
and 15.0,
inclusive.
This column may be NULL if the software did not
report this value.
A time stamp indicating the minute in which the maximum wind speed[166] was recorded.
This column may be NULL if the software did not
report this value.
Atmospheric pressure at the TimeStamp, expressed in kPa
and corrected for elevation. Standard atmospheric pressure
at sea level is 101.325 kPa, so this column's values must
be between 96.3 and
106.3, inclusive.
This column may be NULL if the software did not
report this value or the reported value was subsequently
recognized as erroneous.
The amount of rain measured since the beginning of
the year, in millimeters. Values must be integers greater
than or equal to
0.
This column may be NULL if the device did not
report this value.
TimeStampRain (Rainfall for this TimeStamp)
The amount of rain that was measured at this WStation since the previous TimeStamp.
This column may not be NULL.
An integer, indicating the number of lightning strikes recorded throughout the hour represented by this row.
This column may be NULL if the software or device
did not report this value.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
The WREADINGS table contains one row for each time a person has collected data from the meteorological instruments. So, each WREADINGS row should have at least one associated RAINGAUGES, TEMPMINS, or TEMPMAXS row, but no more than one associated row from any one of these tables.
Note
Automated weather readings are not recorded in WREADINGS .
For any one weather reading the minimum recorded temperature cannot exceed the maximum recorded temperature -- the TEMPMINS.Tempmin value related to the WREADINGS row cannot exceed the related TEMPMAXS.Tempmax value.
The combination of WRdaytime and Wstation must be unique.
The Wstation column cannot be changed when there is a related RAINGAUGES row.
A unique positive integer representing the meteorological data collection event.
This column is automatically maintained by the
database, cannot be changed, and must not be NULL.
Code indicating the station from which the data were collected. Must be a value on the WSTATIONS table.
The day and time the meteorological data were collected. The time zone is Nairobi local time.
TRUE when the WRdaytime
column contains an estimated time. FALSE when the WRdaytime column is an accurate record
of the time the measurement was taken.
Initials of the person who collected the data. Must be a value contained in the Initials column of a row on the OBSERVERS table.
Textual notes on the weather reading.
This column may be NULL when there are no
notes.
This column may not be empty, it must contain characters, and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
Tables in this section contain data about data, but they are not support tables. These tables are broadly applicable to many different kinds of data, and thus do not belong in any other section.
This table lists and explains "behavior gaps": periods of time during which data (e.g. censuses, interactions, focal sampling) for an indicated group are (or are suspected to be) sparse, lacking, or simply lower than normal, for a known reason. The "known reason" is an important element of these gaps; periods of time where data collection happens to dip below the norm for unknown reasons are not included in this table.
Data from gap periods are not any less "valid" than data from any other times. However, when aggregating and analyzing data, the sparseness of data in a given period may affect the final results. The purpose of this table is to point out such periods and allow users to decide for themselves how to deal with them.
Reasons for gaps vary widely, so they are noted in a text column rather than with a support table of possible "gap reasons". This makes querying for reasons unwieldy, but this is by design; the table is intended to be used as a guide for thoughtful consideration[167] of time periods where gaps in observation may be affecting analyses.
When discussed in this table, a "gap" does not necessarily mean a complete absence of data for the indicated period. It may merely refer to periods where collected data is sparser than usual. Also, a gap does not necessarily indicate that all data types are uniformly sparse. It may be that the gap only applies to a single type of data. Users should pay attention to the Gap_End_Status and Notes columns for details about which data types are affected.
Identification of a gap is done by a data manager. The system is not involved with this process, and does not handle data from gap periods differently than data from any other time periods. Those kinds of judgments are left for the user to make.
A group may have overlapping behavior gaps; it's possible for more than one factor to affect observation of a group at the same time.
A gap's Gap_End must be
after its Gap_Start, or NULL.
The Gap_End can only be NULL
if the group's GROUPS.Cease_To_Exist is NULL. This allows for
recording of ongoing, not-yet-completed gaps.
A gap's Gap_End and Gap_End_Status must both be NULL or
both be non-NULL.
A unique integer identifying the BEHAVE_GAPS row.
This column is automatically maintained by the
database and must not be NULL.
The Gid of the group affected by this gap.
This column must contain a Gid value of a row on the GROUPS table. This column may not be NULL.
The date on which the gap began. This date must be between the group's GROUPS.Start and GROUPS.Cease_To_Exist, inclusive.
This column may not be NULL.
The date on which the gap ended. This date must be between the group's GROUPS. Start and GROUPS.Cease_To_Exist, inclusive.
This column may be NULL, see above.
The reason for, or status of, the gap's end. The legal values for this column are defined by the GAP_END_STATUSES support table.
This column may be NULL, see above.
Text notes about the gap, especially information about the gap's cause.
This column may not be empty, it must contain characters, and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
[26] There are, of course, also system generated row identifiers, which are arbitrary and not derived from any field collected data.
[27] This is a generated error instead of one that is immediately raised in order to ease the data entry process. Because births are recorded before CENSUS rows are entered so that new births do not raise errors when uploading census data, new births regularly have dates that follow the mother's Statdate. This could be avoided by entering births without a Pid and then updating the Pid once the CENSUS table has been updated but this was deemed overly burdensome.
[28] Recall that when an individual has no non-absent CENSUS rows, their Statdate is set to their Entrydate, which might be before the LatestBirth. It is therefore presumed that a Statdate being before a LatestBirth will only ever be a temporary occurrence that will go away after the individual's CENSUS data have been added.
[30] This number was chosen based on data management
minutiae related to the fact that a single census in a group
can interpolate an individual present in the group for up to
14 days. If this
value in the interpolation code ever changes, then the
number of days that LatestBirth
is allowed to be after Entrydate
should be re-evaluated.
[31] Thanks to the annoying habit of certain months to not be exactly thirty days — not to mention "leap days" — it's possible that different users may have slightly different interpretations of how many days are contained in "X" years. To allow some flexibility when making these estimates, this rule is implemented as a warning and not an error.
[32] This column was added when PostgreSQL depreciated its “hidden” identifier column, Oid.
[33] This is unlikely as the database will not allow entry of a duplicate Sname.
[34] At the time of this writing, the focal sample
data collection devices use the Sname
XXX for their own special purposes.
There may be other such reserved Sname values unknown
to Babase.
[35] Or whatever you want to call it in the case of a fetal loss.
[36] This is termed a visit in the Protocol for Data Management: Amboseli Baboon Project, which should be consulted for further details.
[38] D usually
occurs when a male is seen alone or in a
non-census group.
[39] When the Status
column is D, the
value of the Cen column
indicates whether or not the individual was
marked “absent” on the field census
for the day.
[40] Facilities exist to require such CENSUS
rows and their associated DEMOG rows be entered in a single
transaction, and the rule requiring CENSUS
rows with a Status of
D to have a
related DEMOG rows could
then be enforced.
[41] DEMOG nearly makes the
M CENSUS
Status code obsolete, were it not
so hard to search on textual data. Indeed, it was created
in response to difficulties with the
M code.
[42] It may seem odd that the Comment column may be
NULL given that this is the only column in the table
containing baboon-related data. However the data
entered into the database can be an abbreviated version
of the actual demography note, abbreviated even into
non-existence.
[43] The system checks the group in which the individual was last censused “present” rather than the individual's Matgrp in order to accommodate group splitting.
[44] Presently group 9.0.
This value is hardcoded
at present.
Individuals are generally put in the unknown group when interpolation does not know their group membership, but it is also possible for an individual to be explicitly placed in the unknown group.
[45] This implies that a GROUPS row's From_group and it's To_group cannot be equal.
[46] As opposed to it being merely a coincidence that the gap began the same date that the group did.
[47] Again, as opposed to it being coincidence that gap and group ended at the same time.
[48] Because there is not a separate column for fusion start date Babase can only track fusions when all groups involved start fusing on the same date. Babase cannot track fusions involving more than one group when 2 groups begin to fuse and others fuse later before the first 2 groups complete fusing.
[49] The precise definition of an "official" study group is left for data management to determine.
[50] In constrast to birth and death, which mercifully tend to be pretty definite.
[51] At the time of this writing, the date used in the case where the transition to sexual maturity was not observed is the date when the individual first came under observation and was already mature.
[52] The “ON” date MSTATUSES code is a special value. See MSTATUSES: Special Values.
[53] Note that this is not literally true, because testicular changes in males are not tracked on a daily basis - males are assigned a matured date on the first day of the month in which seen with fully round testes. Likewise, a female's first Tdate will sometimes have a few days of error around it, as might other transitions.
[54] ...or average, or standard deviation, etc....
[55] This value was chosen somewhat arbitrarily. It's certainly possible to have more than 9 of a particular wound or pathology affecting a body part, but for our uses such a high number is implausible. This value may need to be adjusted in the future.
[56] Therefore during periods of continuous observation no sexual cycle transition events can go unrecorded. See the CYCPOINTS documentation below for the constraints this places on CYCPOINTS within a series.
[57] Yes, updates to CYCPOINTS can result in automatic changes to the CYCGAPS.State, meaning that updates to both tables are occurring in a single transaction. This is okay, because updates to State do not result in changes to CYCGAPDAYS.
[58] Admittedly, validation on CYCPOINTS and other tables could be rewritten to eschew CYCGAPDAYS and use CYCGAPS instead. However, that would result in a major performance dip, so let's not do it unless we have to.
[61] See Appendix C for an example.
[62] This rule minimizes the degree to which CYCPOINTS move between cycles, minimizes the degree to which their Cids change.
[63] It may not be worth documenting this, as there are certainly cases where it is not clear which rows are “earlier”. One such case is changing the date of a Ddate to a later date, that fall after subsequent cycles. If there is concern about the permanence of Cids then it may be best to simply delete CYCPOINTS rows and re-insert them rather than modify existing rows. This at least gives the greatest degree of control over the Cid values.
[64] Quite a bit of Babase's logic relies on there being a continuous series of Mdate, Tdate, Ddate sequences unless there are gaps in observation. It is for this reason that cycles must be “complete”.
[65] This is checked rather than enforced by index or trigger because the condition must exist temporarily as the triggers update the Seq.
[67] The system allows the condition to occur to provide an opportunity to insert a new Mdate, Ddate, Tdate aggregate -- a new cycle -- into the middle of a period of observation. One of these dates must be inserted first, breaking, for the moment, the pattern of cycling -- the repetition of the Mdate, Ddate, Tdate sequence.
[68] This is enforced in triggers rather than by index as the triggers use this condition as a test for whether a new CYCLES row must be created.
[69] It is expected that such rows will exist only until PREGS.Conceive is updated with a reference to them.
[70] Note that cycles may be “cut off”, for a variety of reasons; some cycles may only contain a single CYCPOINTS row, that is, the Cid value may be unique to a single CYCPOINTS row.
[72] Or was in progress when observation ceased, which Babase treats the same as pregnancies in progress at the time data entry ceased. When “now is” is an important consideration in the determination of what in progress means. The cessation of data entry (e.g. BIOGRAPH.Statdate), for whatever reason, is the closest Babase comes to the concept of “now”.
[73] This implies that each Resume value differs from all the others.
[74] Zdate really.
[75] This condition also ensures that a female will not have more than one ongoing pregnancy, as pregnancies require a conception cycle.
[76] It is expected that such Tdates will exist only long enough to update a pregnancy's Resume value.
[77] There should only be CYCGAPS rows when a sexual cycle event may have been missed, but clearly when there is a CYCPOINTS.Resume value then no sexual cycle was missed.
[78] The MATERNITIES view does exactly this. It can be used whenever there is a need for these tables to be joined in this way.
[79] Why is this round-about-the-barn way preferred? Because curmudgeonly old database designers like to insist that keys contain no meaningful information, that's why.
[80] See? We told you that keys should not contain meaningful information.
[81] This indication of a period of no observation is not validated against the CYCGAPS table, that serves as a record of periods of no observation which are long enough that a sexual cycle transition event (Mdate, Tdate, or Ddate) may be missed. Babase does not have records of periods of no observation that are long enough to miss pregnancies. Although it would seem that CYCGAPS could be used for this purpose, and indeed CYCGAPS does “black out” REPSTATS, validating parity against CYCGAPS has not been thought through and awaits a future Babase enhancement.
Regardless, Babase does not presently automatically place a parity in the 100's -- the decision to switch between the 100s and the 1s (or 10s) must be made manually.
[82] This criteria is carefully phrased to account for gaps in the recorded data during the time period in which deturgesence probably began.
[83] When an individual matures, at menarche, there is no Mdate in the first sexual cycle.
[84] notably consortships
[85] There is no restriction on the age or maturity status of the female.
[86] This is not always as useful as it seems. See the rationale for the PARTS table.
[87] It is not that these interactions never occur among young individuals, it is that the researchers' interest is in paternity and maternity and so find that having to concern themselves with filtering out sexual interactions between juvenile individuals is distracting.
[89] and perhaps ejaculation
[90] Presumably data that is collected on a Psion or other electronic device.
[91] Requiring INTERACT_DATA.Observer be NULL, even when
the existing value is “correct” and
synchronized with SAMPLES.Observer, ensures that the value of
the observer column has been taken into consideration by
the person modifying the database.
[92] Consult the Amboseli Baboon Research Project Monitoring Guide to be sure, but this is because the accuracy of the data are never more than one minute, if that.
[93] See the appendix: The All-Occurrences Focal Point Data.
[94] As opposed to recording the interaction with an electronic device.
[95] Whether or not a MPI_DATA row records a request for
help is determined by whether or not the value of the
related MPIACTS.Kind column is
R.
[96] Because the individual from whom help was requested is unknown, there is no way to tell if help was given in response to the request.
[97] Note that if we had the time the sample started, to the second, and we knew that the operator never took more than 59 seconds to enter the point data, and we assume that the operator makes the observation when the timer chimes, then we could calculate the actual time the point was observed. Absent these conditions it appears difficult or impossible to tell which of the 1 minute observation intervals were missed when there is not an exact match between the number of points taken and the total number of minutes in the sample.
[98] It is possible to create a view that extends the
NEIGHBORS table by adding another column, call it
Neighbor, that contains either the Sname or the Unksname,
which ever is not NULL. However, the utility of such a
column is not obvious because it seems that any analysis
done using such a column would have to consistently use
outer joins and then constantly test for NULL results,
lest the Unksname data disappear from the analysis. At
first glance this seems similar to the testing which must
be done to when using two separate columns, the existing
design, so it is not clear whether there's anything to be
gained.
Such a view can always be added in the future without breaking backward compatibility.
[100] The information on the actual unknown neighbor codes used in the field does not appear to be in the Amboseli Baboon Research Project Monitoring Guide.
[101] The name of the focal individual is always recorded, as there is always the intention to observe the focal individual even though this does not always happen.
[102] As the values in the POINT_DATA.Ptime column has little to do with the actual time of observation, it is impossible for Babase to perform additional consistency checks to between the points and the corresponding summary information in SAMPLES. Fortunately, as the data loading process is automated, there is little opportunity for data corruption.
[103] As all observation occurs during the day there are no issues surrounding samples taken just before midnight that start on one day and end on the next. Should there ever be such, this should be the date the sample started.
[104] The anesthetic administration times are not aggregated in this view although it could be useful to aggregate the difference between the time of darting and the time additional anesthetic was administered.
[105] To cover the case where Dartings-Pickuptime is
NULL.
[107] The column is allowed to be NULL due to data
entry procedural constraints. The first data uploaded
creates rows in DARTINGS but the data
set containing mass is not uploaded until later.
[108] In a canonical database design this column would be on the DPHYS table. The column is part of the DARTINGS table due to concerns that the column might be overlooked by a user because so many other note columns are on the DARTINGS table.
[109] In a canonical database design this column would be on the DART_SAMPLES table. The column is part of the DARTINGS table due to concerns that the column might be overlooked by a user because so many other columns are on the DART_SAMPLES table and DSAMPLES view.
[110] This behavior exists so that rows can be inserted into TEETH via the DENT_CODES view.
[111] The alternative to this, an approach closer to the “ideal” database design, is to have separate tables for width and length measurements. This seems excessive.
[112] This rule is a result of the aforementioned design choice that places Testwidth and Testlength in the same table. A consequence of this choice is that this rule must exist to ensure that Testseq values are, effectively, contiguous.
Note that this condition must remain true even while
the rows are in the process of automatic
re-sequencing. It may be that some combinations of
data values will simply not work with all possible
UPDATE statements that change the row
sequencing. Those experiencing problems should delete the
rows in question and re-insert them with the correct
sequence numbers.
[113] The alternative to this, an approach closer to the “ideal” database design, is to have separate tables for width and length measurements. This seems excessive.
[114] This rule is a result of the aforementioned design choice that places Testwidth and Testlength in the same table. A consequence of this choice is that this rule must exist to ensure that Testseq values are, effectively, contiguous.
Note that this condition must remain true even while
the rows are in the process of automatic
re-sequencing. It may be that some combinations of
data values will simply not work with all possible
UPDATE statements that change the row
sequencing. Those experiencing problems should delete the
rows in question and re-insert them with the correct
sequence numbers.
[115] Also "tissue" and "tissue sample", but those two terms aren't terribly different anyway.
[116] Even in the coldest of cold storage, frozen samples will slowly evaporate over time. A 100-μL sample that is frozen and stored for 5 years is unlikely to still be the full 100 μL at the end of that time.
[117] It is presumed that any reader who cares enough about nucleic acid samples to read this documentation is already familiar with the polymerase chain reaction. We will not attempt to explain it here.
[118] Admittedly, this approach is imperfect and is likely underestimating the true prevalance of the problem. The date written on a sample may not be the true date it was collected but may still be a date that the individual was censused. Unfortunately, there is little else that the system can do to recognize when this occurs.
[119] That is, the population whose data are recorded throughout the many tables in Babase.
[120] Related rows in this table are automatically inserted when rows are inserted into BIOGRAPH, so manual insertion of these rows is effectively not allowed.
[121] Similar to inserts, related rows in this table are automatically deleted when rows are deleted from BIOGRAPH, so manual deletion of these rows is effectively not allowed.
[122] Waterholes may be more or less permanent features of the landscape, or only temporary rain pools. This is no surprise to those familiar to the SWERB dataset, but whenever waterholes are mentioned in relation to SWERB data the “waterhole” may be either a waterhole or a rainpool.
[123] It is believed but not certain that this is the way PDOP is used.
[124] It is not clear whether the accuracy is 2 or 3 dimensional vector; whether the reported distance includes error in altitude.
[125] Because database rules which enforce when PDOP and
Accuracy values must be NULL are hardcoded into the
database it will take programmatic changes to change these
limits. Normally this would be avoided by adding a column
to the GPS_UNITS table to indicate whether
or not the particular GPS unit records a PDOP or accuracy
reading, thus allowing new units to be introduced which
record such data. However because records have been lost as
to which specific GPS units were used when and, as of the
time of this writing, no one wishes to reconstruct the
categories of GPS units in use based on a PDOP/Accuracy
capability criteria the system design uses hardcoded dates
to validate. Note further that given the existing set of
validation criteria for PDOP and Accuracy there is never a
circumstance which requires a PDOP or accuracy to be
present. Normally the values of GPS_UNITS.Errortype
would force the presence of PDOP or Accuracy values.
Instead they merely enforce their absence. This is partly
for reasons similar to the preceding and partly because,
particularly during periods when GPS data was
hand-transcribed, sometimes data is missing.
[126] And, possibly, subsequently corrected by the data specialists after consultation with the field teams.
Because the data manager expands the observer codes in the departure rows from 1 to 3 characters the SWERB_DEPARTS_GPS.Garmincode column can hold more than 10 characters.
[127] From a database design perspective it would make sense to control whether or not a Garmincode must be present based on a column in the GPS_UNITS table. In practice because all future GPS units will very likely allow the entry of data when waypoints are taken the matter is moot.
[128] While it may be desirable to have a cutoff date after which all data obtained using GPS units must come from the GPS units themselves, no such cutoff date has been established.
[129] Electronic manufacturers have taken to silently changing the specifications of a device without changing the model, a situation which is quite annoying when the specifications matter. When no other sort of identifying information is available sometimes the serial number can be used to determine device capabilities.
[130] The Amboseli Baboon project data protocols require these codes have a particular structure. Babase does not enforce these requirements, primarily because the QUAD_DATA table is essentially a support table and, once created, is static so enforcing specific rules in the database is not worth the time.
[131] Note that rows that violate this rule are not instantly rejected; the error is caught at the time of transaction commit. This is so that during data entry Btimeest and Etimeest values may be entered without Start and Stop values in the expectation that by the time the transaction is committed the insertion of SWERB_DATA rows will have automatically filled in the missing Start and Stop values.
[132] This last check is also performed at transaction commit time, for the same reason.
[133] Ideally, a begin or end time should not be NULL
unless the records have been perused and no time found, in
which case the time source would always be
bb_norecord when there was no time. In
practice this has not been done.
[134] Note that this rule is tested for immediately, not at
the time of transaction commit. This
means that the Btimeest and
Etimeest columns must be
non-NULL before inserting SWERB begin and end rows that
have non-NULL times.
[135] More precisely, when the SWERB_BES.Seq is
NULL. This typically amounts to the automatic
sequencing of newly inserted rows because those are the
rows which typically have no Seq value.
[136] At first glance it would seem appropriate to
sequence those SWERB_BES rows with
NULL Start times based on
the first related SWERB_DATA.Time value but this presents a
number of problems. Such a design would not allow for any
flexibility in manually re-sequencing such rows unless
automatic sequencing took place only upon insert of SWERB_DATA rows, in which case inserting and
then deleting the inserted row could change the sequencing
of the SWERB_BES rows. Such
un-reversible changes can be confusing.
[138] Manual sequencing is therefore only useful when the
SWERB_BES.Start is NULL or when there are
“ties”. Sequencing is normally manipulated
by changing SWERB_BES.Start values, which are themselves
automatically picked up from SWERB_DATA
rows with B Event values.
When testing for correct sequencing of a SWERB_BES row other bouts of observation
(other SWERB_BES rows) related to the
same group on the same day cannot have a smaller Seq and also have a Start value greater than the
smallest related SWERB_DATA.Time related to the given row. In
those cases where other bouts of observations related to
the same group on the same day have a NULL Start value the comparison is
instead against the other bout's earliest related SWERB_DATA.Time
value. SWERB_DATA rows with NULL
Time values are ignored by
the automatic sequencing process.
[139] This can cause indeterminate results when more than one row is changed in a single update statement.
[140] It generally makes sense to use the last created
SWERB_BES.BEId. If a BEId has been created during the
current PostgreSQL session this can be referenced using
the PostgreSQL expression
currval('swerb_bes_beid_seq').
[141] Allowing changes to the SWERB_BES.DId column would make it difficult to maintain the automatic sequencing of the Seq values.
[142] Allowing changes to the SWERB_BES.Focal_grp column would make it difficult to maintain the automatic sequencing of the Seq values.
[143] All the lines of data dumped from the GPS units are represented as rows in the SWERB_DATA table with the exception of the departure records.
[144] When a group has fragmented a fragment of the group other than the focal fragment may be observed at some distance.
[145] For the occasional “unknown other group” sighting.
[146] These rules imply that when a group is in the process of undergoing fission that the data collection team taking SWERB observations will not flag one of the semi-permanent fission group having it's own code in the groups table a “subgroup” -- unless that semi-permanent group has itself temporarily split.
[147] As of this writing, it isn't recorded in Babase at all. This may change in the future.
[148] I.e. guessed.
[149] Although the system design allows SWERB_GWS rows to represent places other than groves and waterholes, at the time of this writing these are the only places recorded -- with the possible exception of rain pools, which count as waterholes.
[150] Otherwise the SWERB_UPLOAD view would not be able to distinguish between the two grove codes, one of them certain, the other a probable sleeping location.
[151] At the time of this writing the only physical landmarks recorded are groves and waterholes/rainpools.
[152] The exception of the unknown group allows for easy creation of bouts of observation of the unknown group. This is useful because all observations, including those of a non-focal group made on an ad-hoc basis, must be made as part of a bout of observation. But such ad-hoc observations of non-focal groups are made, wait for it, on an ad-hoc basis. A bout of observation may not be in progress. The creation of bouts of observation of the unknown group provide a convenient way to ensure such non-focal group observations are part of an observational bout, and hence are related to an observation team's daily effort -- to a SWERB_DEPARTS_DATA row.
[153] See the preceeding footnote for further detail.
[154] Checking ascent into sleeping grove rules at the time
of transaction commit allows
end-of-observation rows that record ascent into a sleeping
grove to be inserted into the database after all other SWERB
rows for that bout of observation. Because sequence
numbering is not related to end of observation and because
of subgroups and because of the possibility of missing end
of observation times (SWERB_BES.Stop may be NULL) it is not always
possible to distinguish the bout of observation which
represents the last observation of the group by the team for
the day without having a bout that is related to ascent into
a sleeping grove. This means that tests related to
end-of-observation cannot be done as rows are
inserted.
[155] At the time of this writing the ADCODES values are structured such that SWERB_LOC_DATA rows that represent the first or
last observation of each group by each observation team on
each day, the rows that record the group's descent from or
ascent into a grove, must have non-NULL ADtime values. The obverse is
also true; SWERB_LOC_DATA rows that are not the first or
last for the team for the group for the day, that are not
associated with the group's descent from or ascent into a
sleeping grove, must have NULL ADtime values.
[156] A similar rule for the end of observation is not feasible. There are time when, after the last bout of observation of the day has ended, the observation team remains in the field and happens to notice and record ascent into a sleeping grove.
[157] This allows the data that is entered in the field as two separate GPS waypoints but which comprises a single SWERB_LOC_DATA row to be inserted into the database piecemeal.
[158] The decision to create the ADCODES table instead of hardcoding values in the SWERB_LOC_DATA.ADcode column is somewhat arbitrary. At the time of this writing the SWERB_LOC_DATA table is only used to relate baboon groups with sleeping groves at the time of ascent or descent, or to relate the groups with waterholes when drinking. Baboons are never related to groves or waterholes for any other reason, nor are baboons ever related to any other landscape feature. Consequently the expectation is that there will be 3 rows created in the ADCODES table, one for ascent, one for decent, and one for neither that is used when groups drink at waterholes -- and that the ADCODES table will subsequently be forgotten.
Never the less, there is little if any extra technical work involved in having an ADCODES table and its presence opens up future opportunities for recording additional relationships between baboon groups and landscape features, opportunities that do not require any additional programming or other technical involvement. It is for these reasons that the choice was made to have an ADCODES table.
[159] Although the Amboseli Baboon Research Project Monitoring Guide has no provision for uncertainty with respect to any location other than sleeping groves the database contains no rules prohibiting such use. Because the SWERB_UPLOAD will not indicate uncertainty unless a sleeping grove is involved having such a rule seems unnecessary.
[160] SWERB_LOC_DATA is itself an extension of SWERB_DATA
[161] Data that are known to be incorrect should not be retained at all.
[162] One would think that the TEMPMINS and TEMPMAXS tables would need a "span" column similar to RAINGAUGES.RGspan, and a table to correspond to RGSETUPS. As it happens the extraordinary diligence of the field staff in taking regular temperature measurements, in conjunction with the keen analytical skills of the Babase user population, make such an enhancement a flagrant extravagance. Or, to put it another way, it mostly works the way it is so we're leaving well enough alone.
[163] 09 Sep 2010 14:08 EDT, from Dion Almond, “Yes all sensors should be good to 1 decimal place”.
[164] This value was provided by Campbell Scientific's PC400 datalogger support software. Whether or not this is the "right" value to use is probably a question for that meteorologist we just told you to ask.
[165] Admittedly, this inability or unwillingness to report hourly rain may just be unique to the WeatherHawk devices.
[166] Either WindSpeed_Max_Km_Hr or WindSpeed_Max_M_S.
[167] As opposed to using a query to let the database do all the considering for you.
Table of Contents
- Darting
- Group Membership and Life Events
- Physical Traits
- HORMONE_KITS (KITS used to assay HORMONE concentration)
- HORMONE_PREP_DATA (Lab PREParations performed on hormone samples)
- HORMONE_PREP_SERIES (SERIES of Lab PREParations performed on hormone samples)
- HORMONE_RESULT_DATA (RESULTs of HORMONE concentration assays)
- HORMONE_SAMPLE_DATA (Tissue SAMPLEs used for HORMONE analysis)
- HYBRIDGENE_ANALYSES
- HYBRIDGENE_SCORES
- HYBRIDMORPH_OBSERVERS (Observers of hybrid morphology scores)
- HYBRIDMORPH_REPORTS (Hybrid morphology score collection events)
- HYBRIDMORPH_SCORE_DATA (Hybrid morphology scores)
- SWERB Data
- Interpolation
- The Social Group Residency Rules
- The Sexual Cycle Day-By-Day Tables
- Sexual Cycle Determination
- Automatic Sequencing
- Automatic Mdate Generation
These tables contain baboon-related data that are the result of analysis. The RANKS table holds the result of a manual analysis, its data may be updated by Babase users. The remaining tables are automatically updated in accordance with changes made to the primary baboon source data. There is no provision for manual modification of the automatically generated tables.
These tables exist because a relatively large amount of effort, either human or machine, is required to populate them. The tables store the results of that effort and make the results readily available to further analysis.
This section first presents the tables themselves. In the case of the automatically populated tables, or whatever portions of the primary source tables are automatically generated, subsequent sub-sections explain exactly how the tables are populated and so provide further insight into their content.
This table records the proportional presence of specific cell types in a blood sample, as determined by flow cytometric analysis. It contains one row for each analysis of each sample; that is, one row per Dartid-Flow_Date pair.
Because of various practical/logistical minutiae, each flow analysis is not connected to a specific sample in the TISSUE_DATA table. Instead, each analysis is connected to the darting from which the blood sample came. It is certainly possible for a blood sample to be analyzed more than once on the same date, but in practice this does not occur. Based on that assumption, each Dartid-Flow_Date pair in this table is presumed to be sufficient to uniquely identify an analysis.
It is unlikely but possible that a sample from the same Dartid will be analyzed more than once. Because of this, the system will return a warning rather than an error when a Dartid appears more than once in this table.
It is beyond the scope of this document to explain how flow cytometry works, how to interpret its data, etc. For this discussion, suffice it to say that cells are treated with fluorescent antibodies that bind specific cell surface antigens, and a flow cytometer measures their fluorescence to determine which cells have which antigens. With this information, various cell types can be identified according to the presence/absence of specific antigens.
Each of the columns representing a specific cell type
— Monocytes, NK, B, Helper_T, and Cytotoxic_T — are percentages
indicating what proportion of the provided sample is comprised
by that cell type. These columns contain the actual
percentage number, not a value that equals the percentage.
For example, 25.00% would be represented as
25.00, not 0.2500.
Because of the implicit relation between these columns’
values, none of them can be NULL and the sum of those
columns must be between 99.9 and
100.1.
The percentages recorded in this table do not represent the percentage of the indicated cell type among all blood cells, nor among all white blood cells. Prior to analysis, peripheral blood mononuclear cells (PBMCs) are purified from a whole blood sample, and it is only those PBMCs that pass through the flow cytometer. Thus, the percentages in this table indicate the proportion of PBMCs that are the indicated cell type.
Note
These analyses are discussed in greater depth in Lea et al 2018, PNAS. Briefly, the cell types are identified as follows:
| Cell Type | Antigens |
|---|---|
| Monocytes | CD3-, CD20+, CD14+ |
| Natural Killer Cells | CD3-, CD20-, CD16+ |
| B cells | CD3-, CD20+ |
| Helper T cells | CD3+, CD8-, CD4+ |
| Cytotoxic T cells | CD3+, CD8+, CD4- |
The date of the analysis — the Flow_Date — must be on or after the date the sample was collected—the related DARTINGS.Date. In practice, it is unlikely but not impossible that the two dates will ever be equal, so the system will return a warning whenever they are.
The antibodies and blood cells used in these analyses are rather labile, such that the accuracy of the analysis suffers if performed too long after the sample is collected and/or after antibody treatment. The system will return a warning when the date of analysis is 3 or more days after the darting date. That is, when the Flow_Date is 3 or more days after the related DARTINGS.Date.
Tip
To identify the individual being analyzed and the sample collection date, see the related DARTINGS.Sname and Date columns.
A unique integer that identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The DARTINGS.Dartid of the darting during which this blood sample was colelcted.
This column may not be NULL.
A number indicating the percentage of PBMCs in this sample that were identified as monocytes.
This column may not be NULL.
A number indicating the percentage of PBMCs in this sample that were identified as natural killer cells.
This column may not be NULL.
A number indicating the percentage of PBMCs in this sample that were identified as B cells.
This column may not be NULL.
A number indicating the percentage of PBMCs in this sample that were identified as helper T cells.
This column may not be NULL.
A number indicating the percentage of PBMCs in this sample that were identified as cytotoxic T cells.
This column may not be NULL.
Comments or miscellaneous information about this analysis.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
Results from white blood cell counting performed on blood smears collected during dartings. Contains one row for each count of a blood smear. Blood smears from a Dartid must first be recorded in the DART_SAMPLES table.
After darting, blood smears are stained using a Giemsa (or similar) stain. This allows for easy identification of different types of white blood cells when viewed under a microscope. The technician systematically scans the slide and counts the number of each cell type present until reaching a high number, usually 100 or 200. The counts are then used to estimate the proportion of each cell type present in the blood.
Occasionally, blood doesn't smear well, and the technician is unable to count even 100 cells before the smear becomes too dense to read. For these cases with lower total counts, users should consider for themselves whether or not enough cells were counted to accurately estimate cell type proportions.
Each row's Count_Date must be on or after the row's related DARTINGS.Date.
The combination of Dartid and Slide_number must be unique.
The Slide_number cannot exceed the number of blood smears recorded in the related DART_SAMPLES.Num.
A unique integer that identifies the cell count.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The DARTINGS.Dartid of the darting from which the counted blood smear was collected.
This column may not be NULL.
The LAB_PERSONNEL.Initials of the person who performed this count.
This column may not be NULL.
An integer indicating which of this Dartid's blood smear slides was counted for this row.
This column may not be NULL.
Comments or miscellaneous information about the counts on this slide.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table records every attempt to determine an individual's paternity. It contains one row per analysis per offspring.
The term, "analysis", is used loosely in this context. The term might refer to a single event one day where a researcher ran a specialized paternity analysis program. It also might refer to a months-long series of software runs and thoughtful inferences. In general, an "analysis" is any instance in which an individual's paternity was investigated and either i) a putative father was identified or ii) researchers concluded that no putative father could be identified. It is left to data managers to decide any further specifics.
An individual cannot be analyzed before they entered the population; the Date must be after the individual's Entrydate.
It is possible that an analysis will identify an individual as the putative father, but for various reasons that individual might not be deemed the "true" father. The system will return a warning when this occurs. That is, the system will return a warning for each Kid-Putative_Dad pair in this table that does not appear as a Kid-Dad_Consensus pair in DADS_CONSENSUS.
The Putative_Dad must be male. Unlike the DADS_CONSENSUS.Dad_Consensus,the system does not enforce any other restrictions on who is allowed to be a Putative_Dad.
A unique integer which identifies the analysis.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The BIOGRAPH.Sname of the individual whose father is investigated in this row.
This column may not be NULL.
The date of the analysis. For analyses that occurred over multiple days, this is the last day of the analysis.
This column may not be NULL.
The BIOGRAPH.Sname of this Kid's father, according to this analysis.
This column may be NULL if the results of this
analysis are inconclusive.
Miscellaneous notes regarding the analysis.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table records each individual's father, based on the consensus of analyses in DADS_ANALYSES. There is one row for each offspring.
Tip
To see both of an individual's parents, use the PARENTS view.
No individual can have more than one dad; the Kid column is unique.
The Dad_Consensus must be male. If the Kid's mother is known, the Dad_Consensus must be alive during the mother's fertile period — his BIOGRAPH.Statdate must be on or after the Kid's conception date (Zdate) minus the 5 day fertile period. Also if the mother is known, the Dad_Consensus must be mature before the conception date — his MATUREDATES.Matured must be before the Zdate. The system will return a warning if the male is not in the mother's supergroup at any time during the fertile period.
A "consensus" dad should be based on one or more analyses. The system will return a warning if a Kid-Dad_Consensus pair in this table does not appear as a Kid-Putative_Dad in DADS_ANALYSES.
The Dad_Consensus may not have been a perfect choice, but merely the best option; for many reasons, the genotypes of the kid, mom, and dad may conflict, or “mismatch”. These mismatches do not mean that the Dad_Consensus is invalid. The reasons for these mismatches are known (e.g. quality of tissue samples, technological limitations) and are considered when doing the paternity analyses. A Dad_Consensus is provided only when the user is reasonably confident of its accuracy, regardless of any mismatches indicated in the Mismatch column.
A kid must have a dad to mismatch with him; the kid's
Mismatch can only be NULL
when the Dad_Consensus is
also NULL.
A Completeness score for this paternity assignment is also given. This score is a categorical expression of how much is known about the genotypes of the kid, mother, and potential dads considered, as well as how much more information is expected to be gained in the future. For example, the Completeness for an offspring with few genotyped potential fathers depends on whether the untyped potential dads are still alive and available for further sampling. If all potential dads are dead, then no further information is likely to arise to inform this assignment and it is probably as “complete” as it will ever be. If several untyped potential dads are still alive, then the assignment has the potential to change in the future and should have a different Completeness score.
Tip
Use the Completeness column when planning a new paternity analysis. It is helpful for determining which paternities should be re-analyzed and which can be omitted from any further analyses.
The BIOGRAPH.Sname of the individual whose father is identified in this row.
This column may not be NULL.
The BIOGRAPH.Sname of this Kid's father, determined from the consensus of all paternity analyses on this individual.
This column may be NULL when a consensus paternity
has not yet been determined for this Kid.
The DADS_MISMATCHES.Mismatch category for the trio of Kid, mom, and Dad_Consensus.
This column may only be NULL when the
Dad_Consensus is also NULL
The DADS_COMPLETENESS.Completeness of this paternity assignment.
This column may not be NULL.
Miscellaneous comments about this consensus paternity or lack thereof.
This column may be NULL This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains all the data supporting the paternity assignment in an analysis, one row per datum.
Each row contains a key-value pair, where the Term is the key and the Datum is the value. For example, if
an analysis determined paternity by comparing individuals'
microsatellite loci, that analysis might have a row in this
table with the Term LOCUS TYPE and Datum
MICROSATELLITES.
The Datum might be a string of text, a number, or a date, but it will all be stored in the Datum column as text. Regardless, the system validates that the Datum is a textualization of the data type intended for the row's Term. For more information see DADS_EVIDENCE_TERMS.Data_Type.
Note
Because the Datum column is used to contain many different kinds of information, validating it is complicated. The warning system can be a helpful tool for enforcing Term-specific validation.
A unique integer which identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The DADS_EVIDENCE_TERMS.DADS_EVIDENCE_TERMS.Term indicating what kind of data is being recorded in this row.
This column may not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
The group membership table. This table records which group each animal is in on which date, excepting fetal losses (individuals with no Sname). There is a row in MEMBERS for every individual for every day between Birth and Statdate, inclusive, including periods during which the whereabouts of an individual are either recorded as being unknown or assumed unknown by the interpolation procedure. (See: the unknown group.) Some living individuals have MEMBERS rows after their Statdate, for more information see the section: Interpolation at the Statdate. MEMBERS is most useful when one is interested in an individual's location on a particular date. Simply check MEMBERS for the individual on that date. To find all the individuals in a group on a date, look at all the rows in the table on that date for the group.
MEMBERS is a single population-wide table created and updated automatically. It contains 3 categories of group membership information on each individual: interpolated physical presence in a group; supergroup, i.e. “origin” group, during periods of fission and fusion; and the more broad residency group social membership category.
Interpolation is designed to
“smooth out” brief periods of no observation. It
guesses which group an individual is likely to be in when
there is no observational data. The interpolated group
membership information is based on information from the CENSUS, BIOGRAPH, and DEMOG tables and stored in the Grp, Origin, and
Interp columns. Interpolation is
described fully in
a section below. The MEMBERS rows which are the result
of guessing have an I as
their Origin value.
Note
Babase requires that an animal be located in exactly one group on any particular day, the combination of Sname and Date should be unique. The intent of this table is to record the location of each animal at the start of each day. See other documents for further information on how the actual practice of data acquisition and entry impacts this goal.
Tip
If your analysis involves group membership and the time period in which you are interested includes a group fission or fusion you may want to be using the Supergroup column rather than the Grp column. An individual's Supergroup does not change until the date fission/fusion completes, whereas Grp fluctuates between daughter/parent groups during periods of fission/fusion. Using Supergroup allows analysis to treat fission/fusion as an instantaneous event rather than one which occurs over time.
The Delayed_Supergroup column is primarily for the system's internal use and can be safely ignored.
Caution
The Supergroup column (and the Delayed_Supergroup column) are not computed automatically. When the CENSUS or DEMOG tables are changed a Data Manager must tell the system to recompute the Supergroup.
Census data, and so MEMBERS.Grp, is expected to record group membership at the most fine-grained level. Normally this directly corresponds to membership in the usual, expected, groups but during periods of group fission or fusion the groups censused may not be actual, permanent, groups. Supergroup information locates an individual within their parent group during periods of fission and fusion. This is stored in the Supergroup column.
An individual cannot be interpolated into a group that
has ceased to exist, or has not yet begun to exist. The Date of interpolated rows — those
whose Origin is
I — must be between
the Grp's related GROUPS.Start and Cease_To_Exist.
Caution
The system enforces this rule "on-commit". In a
transaction ending with a ROLLBACK, any
changes to this table will not be validated against this
rule. This means it is possible for an invalid change to
appear error-free if executed in a rolled-back
transaction. Committed transactions (and commands executed
outside of transactions) perform this check as
expected[168].
The third category, group residency, is designed to reflect social membership within a group. This as opposed to physical presence. The rules for residency are described in the section on group residency. Residency is based on the GROUPS, BIOGRAPH, CENSUS, and DEMOG tables. Residency results are stored in the Residency, LowFrequency, and GrpOfResidency columns.[169]
When Residency is
R, GrpOfResidency may not be NULL.
Otherwise GrpOfResidency must be
NULL.
When residency is assigned, the density of data used to
make that assignment must also be indicated. GrpOfResidency and LowFrequency must both be NULL (when not
resident) or both non-NULL (when resident).
Note
Social group residency may differ from physical group presence -- the GrpOfResidency value may differ from the Grp value. This is particularly true of males who visit other groups.
Caution
Social group residency is not computed automatically. When the CENSUS or DEMOG tables are changed a Data Manager must tell the system to recompute the social group membership.
Babase populates this table automatically. For the most part users cannot directly manipulate the table's data, although the data managers must manually trigger residency analysis.
A unique integer which identifies the MEMBERS row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The individual whose location is being recorded. The three-letter code that identifies the individual's row in the BIOGRAPH table. There will always be a row in BIOGRAPH for the individual identified here.
This column may not be NULL.
The group where the individual is located. This is a Gid value from GROUPS. This field should contain the most specific sub-grouping available -- subject to the constraints of the data entry protocol, of course. Aggregation into larger groupings is accomplished by way of the Supergroup column.
This column may not be NULL.
A one letter code indicating the source of the
location information. This information is derived from,
and has the same values as, the Status column of CENSUS, with the exceptions that
MEMBERS.Origin contains the
I (interpolated) value
not found in CENSUS, and
does not contain the A
(absent) value. The codes are as follows:
C (CENSUS) values represent census
data points, I
(interpolated) values are derived from the census data
points, D (demography) values
represent demography notes not present in the census
sheets, M and
N (manual) values
represent census data points due to operator intervention
in CENSUS . The
S, E,
F, B,
G, T,
L, and R
codes are derived from analysis of historical data. See
the CENSUS section for
further information.
This column may not be NULL.
The time interval, in days, from the date in which
an individual was previously observed to be in a group
(censused or born into group -- automatic placement in
the unknown group does not count) to the date of the
MEMBERS row. So the value
is 0 on those days on which the
individuals are censused (and on the individuals' birth
dates), 1 on those (non-census) days
immediately before or after the census days, etc. For
those MEMBERS rows in which
the interpolation procedure has associated an individual
with the unknown group, for lack of a better place to
put them, the Interp column is the number of days
“distant” from the interpolating CENSUS row, or the birth date,
that determined the group membership. Note that the CENSUS row that determined that
the MEMBERS.Grp should be
unknown may record an absence.
Important
The Interp value is not meaningful over intervals
that contain census rows that are themselves the result
of an analysis. Over these intervals Interp is NULL.
For more information see Interpolation, Data are not Re-Analyzed.
This column many be NULL.
The Gid of the permanent group[170] in which the individual is a member on the given MEMBERS.Date.
Between a group's GROUPS.Permanent date and it's GROUPS.Cease_To_Exist date, inclusive, individuals within the group have a Supergroup value of the group itself.
During fission the supergroup of a fission product on a given date is the parent group, i.e., the GROUPS.From_group.
During fusion the supergroup of a fusion product on a given date is the parent group in which the individual was most recently censused. I.e. one of the GROUPS with a To_group value of the daughter group's Gid, which is also permanent on the given date, i.e. which has a GROUPS.Permanent value on or before the given date and a GROUPS.Cease_To_Exist value on or after the given date, in which the individual was most recently censused. It is an error if no parent group is permanent.
If there is no parent group the group is its own supergroup.
If the group has no Permanent date but its parent group has ceased to exist, the group is its own supergroup. This is the only way that a non-permanent group can be a supergroup.
This column may be NULL when the supergroup has
not yet been computed.
The supergroup, calculated with a delay of 29 days.
This column is used internally by Babase.
This column may be NULL when delayed supergroup
has not yet been computed.
Whether or not the individual is resident in the group on the given day. The legal values for this column are:
| Code | Description |
|---|---|
R | Resident -- in the GrpOfResidency group |
N | Non-Resident -- physically present in a known group, but not a resident anywhere |
U | Unknown -- whereabouts unknown, residency unknown |
X | EXcluded -- this date was excluded from the residency analysis, probably because it is before the individual's Entrydate or after their Statdate. |
This column is NULL when the row's residency has
not yet been computed.
A boolean that indicates if the 29-day "window" that was used to determine this row's GrpOfResidency had sparse census data. This "window" is usually — but not always — this row's Date and the subsequent 28 days. In some cases a different "window" may be used, as discussed in the section on group residency.
When the pertinent 29-day window
has 3 days or fewer on which
census information is recorded — days for which the
individual has rows in CENSUS —
this column is TRUE. The censuses can record absence or
presence.[171] Being interpolated does not count.
If the above criteria are not met for the pertinent
29-day window, this column is
FALSE.
Caution
CENSUS.Status codes other than
C,
D,
M, and
A can often be repeated
continuously over periods of many consecutive days.
This can distort the low frequency determination.
This column may be NULL when residency has not yet
been computed or when the individual is not a resident on
this date.
The social group in which the individual belongs. This is a Gid value from GROUPS.
This column is NULL when residency has not yet
been computed as well as when residency is not
established.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
The ranking of individuals within groups. This table contains a row for every month for every ranked individual for every type of rank assigned to the individual. When the ranking has not been done for a type of rank in a month, there are no rows for members of that group for that month with that type of rank.
Rankings are determined via a manual process that considers both quantitative information, such as the outcome of agonism interactions within a particular month, and some qualitative judgments such as other observed behavior during and surrounding the month in question. As such the rankings are somewhat smoothed and are not strictly dependent upon observations made within a single 1 month time interval. For further information please consult your local Babase scientist.
The system will report a warning when a ranking of some Rnktype has been done on a group and there are individuals (returned by the RNKTYPES.Query) who have not been ranked.
Rankings may be based on irregular observations of a group before the long-term study began, or before it became an "official" study group. Either way, the ranks for such a group will likely be before any of the individuals' Entrydates. Because of this, the system will allow but issue a warning when an individual's Rnkdate is before the first of the month of the individual's LatestBirth, and another warning if before that of the Entrydate.
The combination of Sname, Rnkdate, Grp, and Rnktype must be unique.
Ranks are assigned within groups, so all individuals must be in the group ranked at some point during the month. Specifically, MEMBERS must record that the ranked individual is a member of the group as determined by the Grp column, during the ranked month.
Caution
Be careful when changing group membership or group rankings; the rank will almost certainly change if an individual's group is changed.
Rank assignments should not be interpreted as absolute truth. They are a "best fit" assignment based on the density and volume of available data. Users should remain aware of this and be prepared to make decisions about the accuracy and reliability of individual rows in this table. Three columns are provided to assist users with these decisions: Ags_Density, Ags_Reversals, and Ags_Expected. These columns provide information about the supporting agonism data involving members in this Grp during the month represented by the Rnkdate and who have a RANKS row with this Rnktype.
To fully understand the meaning of these columns, it is helpful to visualize the supporting agonism data in a matrix. Across the top of the matrix, all ranked individuals for a given Grp, Rnkdate, and Rnktype are listed in numerical Rank order (rank #1 is left-most). On the left side of the matrix, the same individuals are listed again in the same order, top to bottom. In each cell of the matrix, there is a number indicating the number of agonisms (from INTERACT_DATA and PARTS) in which the individual on the x axis was submissive to the individual on the y axis. In other words, it is the number of times that the individual on the y axis "won" over the individual on the x axis. See the example matrix below.
Example 4.1. An Agonism Matrix
For a given Grp and specific Rnkdate and Rnktype, suppose that there are only four individuals named ABC, DEF, GHI, and JKL, ranked in that order. A matrix of the agonisms between these individuals in the relevant time period might look like this:
-----|-ABC-|-DEF-|-GHI-|-JKL-
-ABC-|-----|--3--|--0--|--2--
-DEF-|--1--|-----|--1--|--1--
-GHI-|--0--|--0--|-----|--5--
-JKL-|--0--|--0--|--0--|-----
As shown here, ABC "won" over DEF 3 times, while in the same period DEF "won" over ABC only once. JKL "won" over no one and "lost" twice to ABC, once to DEF, and five times to GHI. The top-left-to-lower-right diagonal is empty, because it is doesn't make sense to "win" against oneself.
Ideally, when the hierarchy is completely linear, the matrix for this Grp-Rnkdate-Rnktype will have 1) only nonzero values above the diagonal, and 2) only zeroes below the diagonal. When this is not possible, the number of agonisms for each dyad above the diagonal will generally be greater than or equal to that dyad's value below the diagonal. For example, DEF will not typically be ranked above ABC because if so, the number of agonisms above the diagonal (1) would be higher than the number of agonisms below it (3).
For more details about how ranks are assigned, see the data management protocols on the Data Management page of the Babase Wiki.
When data are sparse, a large number of dyads in an
agonism matrix will have zero agonisms above the diagonal.
When data are dense, relatively few dyads above the diagonal
will be zero. The Ags_Density is a
proportion that shows the density/sparsity of the related
agonism data. In an agonism matrix for all individuals with
the same Grp-Rnkdate-Rnktype, the
number of dyads in the top half with a nonzero value is
divided by the total number of dyads in
that matrix's top half. This value is the Ags_Density for all rows with that Grp-Rnkdate-Rnktype. For the rare event that there is
only one ranked individual and therefore no dyads, the special
value 99 is used
instead. With the exception of this one special case, higher
values in this column indicate that a larger number of dyads
had observed interactions that month, so the rankings are
based on a relatively large amount of information. In
contrast, lower values indicate that a smaller number of dyads
had observed interactions, so the rankings are based on less
information.
Note
Historically, the average Ags_Density for all baboon project ranks
is roughly 0.3. While the theoretical
maximum Ags_Density value is
1, in reality this occurs very rarely and
only in small groups. It is especially difficult to attain
high Ags_Density values in larger
groups, because there are more dyads needing data.
It is possible to "win" over an individual but still be
ranked below them, as shown above. The Ags_Reversals column indicates how many of
these so-called "reversals" that the individual experienced.
That is, the number of this individual's agonisms — both
wins and losses — that are below
the diagonal on the matrix for this Grp-Rnkdate-Rnktype. The Ags_Expected column shows the opposite: the
number of this individual's agonisms — both wins and
losses — that are "expected" because they are
above the diagonal. When there is only
one ranked individual and thus no dyads, both of these
columns' values will be 0.
Note
The Ags_Density is a proportion of the number of dyads, while the Ags_Reversals and Ags_Expected are sums of the number of agonisms.
The Ags_Density, Ags_Reversals, and Ags_Expected values should not be calculated
immediately after a row is added, because their values depend
on knowing all of the ranks for the Grp-Rnkdate-Rnktype[172]. Because of this, these columns can be NULL.
Their values are calculated by the rebuild_ranks() function, which should be manually
executed soon after new RANKS rows are
inserted.
The system will return a warning for any row in RANKS with a NULL Ags_Density, Ags_Reversals, or Ags_Expected.
Tip
You may want to use the PROPORTIONAL_RANKS view instead of this table. It includes all of the same columns as this table, but also calculates the ranked individual's "proportional" rank.
A unique integer which identifies the RANKS row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The individual whose rank is being recorded. The
three-letter code which uniquely identifies an individual
(in Sname) in BIOGRAPH. There must always be a
row in BIOGRAPH for the
individual identified here. This column must not be
NULL.
A date that falls on the first day of a month,
representing The year and month of the ranking. The year
must be between 1940
and 2040,
inclusive. This column must not be NULL.
Tip
Use the rnkdate() function to obtain the first day of the month when writing queries.
The kind of rank assigned to the individual, a
Rnktype value from the RNKTYPES table. This column may
not be NULL. Examples of various rankings are: Adult
Females, All Females, etc., as defined in the RNKTYPES table.
This is the ranking among all the animals of the
Rnktype in the group over the Rnkdate period. The most
dominant individual is given a rank of
1, the next most dominant a rank of
2, etc. This information is updated
through the ranking program and as a rule need not be
manually updated. This column must not be NULL. The rank
values must be contiguous and start with
1.[173]
For this Grp-Rnkdate-Rnktype, the number of dyads with observed agonisms in the "expected" direction divided by the number of possible dyads with agonisms in the "expected" direction.
This column may be NULL, but only while awaiting
the insert of all rows from this Grp-Rnkdate-Rnktype.
For this Grp-Rnkdate-Rnktype, the number of agonisms that this individual experienced that were "reversals".
This column may be NULL, but only while awaiting
the insert of all rows from this Grp-Rnkdate-Rnktype.
For this Grp-Rnkdate-Rnktype, the number of agonisms that this individual experienced that were in the "expected" direction.
This column may be NULL, but only while awaiting
the insert of all rows from this Grp-Rnkdate-Rnktype.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table records periods (or “bouts”) of time where an individual remained resident in a group. While residency is shown in MEMBERS on a daily basis, in this table those data are condensed into discrete “bouts”: one for each row. This table also includes information showing why each bout started and finished, and how often observation of the individual was designated "low frequency".
A bout of residency is a period of time in which the individual is resident in the same group on every constituent date (every row in MEMBERS). The individual may be present elsewhere during this time, but the group in which they were resident cannot change, as discussed in the Residency Rules. A change in MEMBERS.GrpOfResidency due to a group fission or fusion is not treated as a group change when grouping residency into bouts.
Residencies may begin and end for reasons that are not immediately apparent. To clarify this, these reasons are indicated in the Start_Status and Finish_Status columns.
For each bout, the prevalence of "low frequency" days
— MEMBERS rows whose LowFrequency is TRUE is provided. This
is shown as a simple count of the number of low frequency days
(Days_LowFreq) and as a
proportion of all days in the bout (Prop_LowFreq).
Note
When considering the prevalence of low frequency days in a bout, both the count and the proportion should be considered. When a bout is especially long, a large number of low frequency days may be obscured when represented as a small proportion. Similarly, when a bout is short, a small number of low frequency days might be magnified when represented as a large proportion.
Tip
To determine the total number of days in a bout, use Finish_Date − Start_Date +1. It is easy to forget the "+1".
The contents of this table are completely dependent on the data in MEMBERS. Data in this table are automatically updated by the system when an individual’s residency data are updated by the rebuild_residency() or rebuild_all_residency() functions. Manual inserts, updates, and deletes can only be performed by an admin.
A unique identifier for the row. This is an automatically generated sequential number that uniquely identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
How or why this residency began. The legal values for this column are:
| Code | Description |
|---|---|
E | Entry — this date is when the individual was first seen, i.e. this is the individual's BIOGRAPH.Entrydate. |
I | In-migration — the individual joined the group after being in another (known) group, i.e. this is not the Entrydate. |
This column may not be NULL.
How or why this residency ended. The legal values for this column are:
| Code | Description |
|---|---|
S | Statdate — this is the last date that the individual was seen, i.e. this is the individual's BIOGRAPH.Statdate. |
O | Out-migration — the individual left this group and moved to another (known) group, i.e. this is not the Statdate. |
This column may not be NULL.
The number of days in this bout that were determined
to be "low frequency" when the individual's residency was
calculated. That is, the number of this individual's
MEMBERS rows between this bout's Start_Date and Finish_Date (inclusive) whose
LowFrequency is TRUE.
This column may not be NULL.
The proportion of this bout's days that were determined to be "low frequency" when the individual's residency was calculated. That is, this bout's Days_LowFreq divided by the number of days in this bout.
This column may not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for each kit or protocol that may be used to assay for the concentration of a hormone in a sample. In addition to general information about the kit or protocol (e.g. the assayed hormone, description/discussion), this table also records information regarding how different kits/protocols for the same hormone can be compared to each other.
The Correction column
indicates how — or if — results from this
kit/protocol should be used with results from other
kits/protocols that assay the same hormone. To do this, it
indicates what adjustments should be made to correct the "raw"
results from this kit. The ESTROGENS, GLUCOCORTICOIDS, HORMONE_RESULTS, PROGESTERONES,
TESTOSTERONES and THYROID_HORMONES views all use this column and the
corrected_hormone() function to calculate a
"corrected" concentration, so all values in this column must
be legal correction inputs
for that function. Specifically: the text in this column must
be readable as a mathematical expression for how the "raw"
value should be adjusted, and when referring to the "raw"
value, the string %s must be used. See
corrected_hormone() for examples
of how these values should be recorded.
The system will return a warning for any rows whose
Correction does not include a
%s.
Tip
When no correction is needed, do not set the Correction column to NULL. "No
correction needed" is indicated with a Correction of
%s.
When the Correction
column is NULL, this is interpreted to mean that it is
unknown how to use or compare the kit's results with data from
other kits. In related views that provide a "corrected"
concentration, this corrected concentration for these results
may be NULL (as in HORMONE_RESULTS), or
the result may be omitted entirely (as in ESTROGENS, GLUCOCORTICOIDS,
etc.).
Example 4.2. Kits with no Correction, and NULL Correction
For a few years, the concentration of "hormone X" was
measured using kits made by the Mojo Jojo corporation. A
few years later, the Mojo Jojo kit was discontinued, and
Hormone X was instead measured using a kit from Utonium,
Incorporated. Analyses suggested that results from the
Utonium kit are more accurate than the Mojo Jojo kit. The
Utonium kit's Correction
would therefore be set to %s, and ideally
the Mojo Jojo kit's data would be corrected to allow
comparison with the Utonium data. However, the actual
measurements from the two kits are highly inconsistent with
each other, so a reliable correction factor for the Mojo
Jojo kit cannot be calculated. Because of this, the Correction column is set to NULL
for the Mojo Jojo kit. In a view showing concentrations of
hormone X (similar to how ESTROGENS shows
concentrations of estrogen), assay results from the Utonium
kit are included, while results from the Mojo Jojo kit are
omitted.
A unique identifier for this kit or protocol. This is an automatically generated sequential number that uniquely identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The HORMONE_IDS.Hormone whose concentration is assayed by this kit or protocol.
This column may not be NULL.
A string of text indicating how to "correct" results from this kit/protocol so that its results may be used alongside results from other kits/protocols that assay the same hormone.
This column may be NULL, when an appropriate
correction has not been determined or is not
possible.
A textual description or discussion about the kit or protocol, including any miscellaneous comments or notes.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every laboratory preparation that was performed on a sample as part of a specific series. Each preparation is recorded with a date, and textual comments may be also be noted.
Tip
Always use the HORMONE_PREPS view in place of this table. It contains additional related columns which may be of interest.
Each row's preparation must have taken place after the hormone sample was freeze-dried and sifted. That is, each row's Procedure_Date cannot be before the related HORMONE_SAMPLE_DATA.FzDried_Date and Sifted_Date columns.
Note
The freeze-drying and sifting of fecal samples that are recorded in HORMONE_SAMPLE_DATA are arguably preparatory procedures. Those preparations are not included here because they affect the whole sample. Recording them in this table would incorrectly indicate that the effect of those preparations is limited to a single series.
The procedure cannot occur more than once in the same series. That is, the Procedure must be unique to the HPSId.
The system will return a warning if an ethanol extraction is recorded in the same series as any other preps[175].
A unique identifier for this preparation. This is an automatically generated sequential number that uniquely identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The HORMONE_PREP_SERIES.HPSId of the series to which this preparation belongs.
This column may not be NULL.
The HORMONE_PREP_PROCEDURES.Procedure performed for this preparation.
This column may not be NULL.
The date on which this preparation finished. If a preparation spans multiple days, then the latest date should be used here.
This column may be NULL, when the date is
unknown.
Comments or miscellaneous information about this preparation.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for each series of laboratory preparations and hormone measurements performed on a specific sample.
It seems worthwhile to use an example to illustrate the meaning of a "series" in this context.
Example 4.3. A familiar "series" of events
One day, Little Miss Muffet decides she wants to have a snack. She gets a bowl of curds and whey, sits down on a tuffet, and proceeds to eat. Soon a spider comes along and sits next to her, frightening her and causing her to run away. Video dramatizations of the event often show her dropping, spilling, or otherwise losing the curds and whey.
Taken together, these events comprise a "series" whose component events could be divided into two groups: preparatory events (get food, sit on tuffet, get scared by spider) and results (run away, lose food). Each preparatory event is preparatory for both of the results, and neither result is dependent or contingent on the other result.
To accurately record these events in a database, Miss Muffet's three preparatory events should be connected to each of the two results, and vice versa. Ideally, separate tables of preparations and results should be in a "one-to-one" or "one-to-many" relationship, but the nature of these data prohibits such an arrangement. This inconvenient "many-to-many" relationship can be addressed by designating each of the many events as components of a single "series".
Similar to Miss Muffet, the process of measuring hormones extracted from a tissue sample is divided into preparatory procedures (in HORMONE_PREP_DATA) and results (in HORMONE_RESULT_DATA), and multiple preparation events may apply to each of multiple results. Their troublesome "many-to-many" relationship is managed by this table, in which related events are grouped into a single series.
This table includes a Series column, which
identifies the series for the sample. The first row for a
sample in this table must have a Series of
1, the second should be
2, and so on. To allow editing or
reordering this value, this rule is only checked after the
current transaction is committed.
A unique identifier for this series. This is an automatically generated sequential number that uniquely identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The HORMONE_SAMPLE_DATA.TId of the sample that was worked on in this series.
This column may not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for the result of every assay performed to measure a hormone concentration in a sample. In addition to the assay's result — the concentration in nanograms of hormone metabolite per gram of fecal sample — each row records the series (from HORMONE_PREP_SERIES) to which this result belongs, the date of the assay, the identity of the "kit" or protocol used to perform the assay, and the mass of sample from which this hormone was extracted. Textual comments may be also be noted.
Tip
There are many views that may be preferred in place of this table. The HORMONE_RESULTS view adds some related columns that make this table more legible for human consumption. Also, there are several views dedicated to specific hormones (ESTROGENS, GLUCOCORTICOIDS, PROGESTERONES, TESTOSTERONES, THYROID_HORMONES) that show assay results with relevant information about the sample and any preps involved with generating the result.
Each row's assay must have taken place after all related preparatory procedures were performed. That is, each row's Assay_Date cannot be before the Procedure_Date of any HORMONE_PREP_DATA rows from the same series (with the same HPSId).
When assay results are generated in the lab, a sample may undergo more than one assay for the same hormone. When there are multiple results, it becomes necessary to determine what the "right" concentration for the sample actually is. Often, it may be best to use the average of all results. In some cases, the results from one kit may be universally preferred over results from another kit. And so on. Laboratory and data managers are presumed to be better-qualified to make such decisions, so those decisions should be made before adding data to this table. To help ensure that this is occurring, a hormone sample cannot have more than one result for each hormone, regardless of the series and the Kit. That is, the combination of the related HORMONE_PREP_SERIES.TId and HORMONE_KITS.Hormone must be unique[176].
The mass of fecal sample that was extracted and measured
in the assay is recorded in the Grams_Used column. This column
should not be NULL. However, in some rare circumstances the
mass can be unknown, in which case the column will be
NULL. Regardless, this is expected to be rare. The system
will return a warning for any assay whose Grams_Used is NULL.
A unique identifier for this assay result. This is an automatically generated sequential number that uniquely identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The HORMONE_PREP_SERIES.HPSId of the series to which this assay result belongs.
This column may not be NULL.
The initial mass of fecal sample from which the assayed hormone was extracted.
This column may be NULL, when the mass is
unknown.
The "raw" concentration of the hormone, in nanograms hormone per gram of fecal sample, determined by this assay.
This column may not be NULL.
Comments or miscellaneous information about this assay.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every tissue sample used for hormone analysis; generally for hormone analysis these will be fecal samples. For each sample, the table records data that are only relevant to hormone analysis and would thus be inappropriate for inclusion in the TISSUE_DATA table.
Tip
Always use the HORMONE_SAMPLES view in place of this table. It contains additional related columns which may be of interest.
For various logistical reasons, it is often not practical for lab personnel to use the database's unique identifier for a sample (its TId) in the lab. Instead, they use a system of their own. The unique identifier used by hormone lab personnel — the "Hormone Sample ID" — is indicated in the HSId column[177].
To analyze a fecal sample for its hormone content, the sample must first be freeze-dried. Following that, the dry sample is sifted into a fine powder, at which point it is ready for whatever preparations are necessary for hormone analysis. The dates that the fecal sample is freeze-dried and sifted are recorded in this table, in the FzDried_Date and Sifted_Date columns, respectively.
A fecal sample cannot be sifted before it is freeze-dried; its Sifted_Date must be on or after its FzDried_Date.
This table attempts to keep an ongoing record of a fecal
sample's remaining mass in the Avail_Mass_g column. It is
left to the user to judge this column's accuracy, which
depends greatly on how diligently the lab personnel keep the
data manager(s) informed of changes. To assist users in making
these judgments, the date that the Avail_Mass_g was last updated
is recorded in the Avail_Date column. A sample's
remaining mass cannot be recorded without also recording this
date; the Avail_Mass_g
and Avail_Date columns
must both be NULL or both non-NULL.
Preparing a fecal sample for hormone extraction and any subsequent handling of the sample must be after the sample was collected. That is, all dates in this table (FzDried_Date, Sifted_Date, Avail_Date) must be after the sample's related TISSUE_DATA.Collection_Date.
The TISSUE_DATA.TId of the tissue sample and the unique identifier for the row.
This column cannot be changed and must not be
NULL.
The "Hormone Sample ID", another unique identifier for this sample. This is a number, created and maintained by lab personnel.
This column may not be NULL.
The date that this sample was freeze-dried.
This column may be NULL, when this date is
unknown.
The date that the freeze-dried sample was sifted.
This column may be NULL, when this date is
unknown.
The mass of this sample, in grams, that is available for use as of the Avail_Date.
This column may be NULL, when the remaining mass
(if any) is unknown.
The date that the Avail_Mass_g was determined.
This column may be NULL, when the remaining mass
(if any) is unknown.
Comments or miscellaneous information about this sample.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
A table listing each analysis that has been performed to generate genetic hybrid scores for individual baboons, with basic information about each analysis.
Each analysis combines statistical techniques with the genetic data available at the time to estimate what proportion of each individual's genome came from ancestry of a specified other species[178]. These estimates are the so-called “hybrid scores”. After several years have elapsed, more individuals are available for scoring, which prompts a new analysis. For many reasons, each analysis may yield somewhat different scores for the same individual. A more-recent analysis does not necessarily negate or supersede an older one, so all analyses are stored here.
The HYBRIDGENE_ANALYSES.Date must be after the BIOGRAPH.Entrydate of all individuals scored in that analysis in HYBRIDGENE_SCORES. Similarly, the system will return a warning for each individual scored in an analysis where the related Date is before the individual's LatestBirth.
A unique integer that identifies the analysis.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The LAB_PERSONNEL.Initials of the person who performed the analysis.
Tip
It's technically possible to have more than one person involved with an analysis, but even in such cases there will certainly be a lead whose initials should fill this column.
This column may not be NULL.
Notes or comments about the analysis.
This column may not be empty, it must contain characters, and it must contain at least one non-whitespace character.
This column may be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
A table listing all the hybrid scores determined by genetic hybridity analyses. Hybridity analyses use statistical tools that may also determine upper and lower confidence intervals[179]. This table also stores those values, if any.
The combination of Sname and HGAId must be unique.
In some analyses, upper and lower confidence intervals
are not generated, in which case the Upper_Conf and Lower_Conf will be NULL. The
system will return a warning in this case.
If either Upper_Conf
or Lower_Conf is provided,
then the other must also; the Upper_Conf and Lower_Conf must both be NULL
or both non-NULL.
When the confidence columns are not NULL, the
individual's Score must be
greater than its Lower_Conf (inclusive), and less
than its Upper_Conf
(inclusive).
A unique integer that identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The HYBRIDGENE_ANALYSES.HGAId of the analysis in which this score was determined.
This column may not be NULL.
The individual's hybrid score for this
analysis. This value must be between 0
and 1 (inclusive).
This column may not be NULL.
The lower confidence interval for the hybrid
score. This value must be between 0 and
1 (inclusive).
This column may be NULL, but only when the
analysis did not generate a lower confidence.
The upper confidence interval for the hybrid
score. This value must be between 0 and
1 (inclusive).
This column may be NULL, but only when the
analysis did not generate a higher confidence.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table records the observers for the hybrid morphology reports, one row for each observer.
A report cannot have the same observer more than once; each HMRId-Observer must be unique.
In practice, an observer does not score an individual more than once on the same date. However, the indicated Date for a score might not be be the actual date of the observation, for reasons discussed below. Because of this, each combination of Observer and related HYBRIDMORPH_REPORTS.Date and Sname should be unique but occasionally will not. The system will return a warning for each non-unique Sname-Date-Observer that appears in these data.
A unique identifier for this observer. This is an automatically generated number that uniquely identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for each time an individual's hybrid morphology is assessed and scored. For details about these scoring events, see the Amboseli Baboon Research Project Monitoring Guide.
Observers do not always record the exact date of the scoring. Instead, they may only record the year and month. When this occurs, the Date is recorded as the first day of the indicated month. It is not possible to discriminate between this and a report that was truly collected on the first day of that month.
An individual cannot be scored before they were first observed, nor after they were last seen alive; the year and month of the Date cannot be before the year and month of the individual's Entrydate, nor can the Date be after the individual's Statdate.
A hybrid score report includes the name of the baboon group in which the scored individual lives. Recorded in the Grp column, this value is provided by the observer(s) and is useful for finding the original paper sheet where the scores were recorded.
Tip
This table's Grp is usually but not necessarily the individual's Grp in MEMBERS on that date. To find the individual's group on the day of scoring, use MEMBERS.
It is difficult to recognize in real time when a group fission/fusion has begun or ended, so the observer(s) might record a Grp that no longer exists. The system will return a warning for any row in this table whose Grp did not exist on the indicated Date.
A unique identifier for this report. This is an automatically generated number that uniquely identifies the row.
This column is automatically maintained by the
database, cannot be changed, and must not be
NULL.
The GROUPS.Gid of the baboon group in which the observer(s) indicated that this scoring occurred.
This column may not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains the hybrid morphology scores that the observer(s) recorded for each of the various traits. Each row is a single score for a single trait.
Tip
Use the HYBRIDMORPH_SCORES view instead of this table. It shows the scores in one row per report, and can also be used to insert, update, or delete data from this table.
An individual cannot be scored more than once at a time for a single trait; every HMRId-Trait must be unique.
Scores are recorded for seven traits: 1) coat color, 2) hair length, 3) body shape, 4) head shape, 5) tail length and thickness, 6) tail bend at the "hook", and 7) muzzle skin. See the Amboseli Baboon Research Project Monitoring Guide for more information.
Which traits are scored depends on the age and sex of the individual. Adults — females age 4 or higher, males age 5 or higher — are scored on all seven traits. Juveniles — females under 4, males under 5 — are only scored on the first five. It is an error to record a score for those last two traits if the scored individual is a juvenile on the related Date. Because the Date may not be exactly right, this rule is enforced with "month"-level precision. E.g. a female cannot have any scores for those last two traits on any date in any month prior to the month in which she becomes 4 years old.
If a report has scored an individual for any trait, there should be scores recorded for all the traits, juveniles and the last two traits notwithstanding. The system will return a warning for any HMRId that does not have scores for all of its expected traits.
The HYBRIDMORPH_REPORTS.HMRId of the report in which this score was recorded.
This column cannot be changed and must not be
NULL.
The trait being scored. This column must be one of the following:
| Code | Meaning |
|---|---|
COLOR | Coat color |
HAIR_LENG | Hair length |
BODY_SHAPE | Body shape |
HEAD_SHAPE | Head shape |
TAIL_LENG | Tail length and thickness |
TAIL_BEND | Tail bend at the "hook" |
MUZZLE | Muzzle skin |
This column cannot be changed and must not be
NULL.
The individual's score for this trait. This number
must be a multiple of 0.5, and must be between
0 and
2.
This column may not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
This table contains one row for every analyzed observation of a location (from SWERB_LOC_DATA).
For a variety of reasons, a GPS point may be collected many meters away from the actual noted location. Because of this, the database does not implement rules that require an observation's GPS coordinates to be within a particular number of meters from the location's SWERB_GW_LOC_DATA.XYSource. Presumably, this means that mistakes that the observers occasionally make go undetected when added to Babase. To help address this problem, a data manager performs periodic analyses of the data and determines scores of "confidence" in the accuracy of the noted location. For further information about these analyses, please consult your local Babase scientist. The SWERB_LOC_DATA_CONFIDENCES table records those confidence scores, and notes the nearest SWERB_GWS.Loc whose coordinates (from SWERB_GW_LOC_DATA.XYSource) are nearest to the related SWERB_DATA.XYLoc and whose SWERB_GWS.Type matches that of the related SWERB_LOC_DATA.Loc.
To determine confidence in a particular observation and to calculate which Loc is nearest, the manager must have a set of "known" or "reference" coordinates. Because of this, the Nearest_Loc should be a Loc value from SWERB_GW_LOC_DATA. The system will return a warning when this is not so -- when a Nearest_Loc value is not a SWERB_GW_LOC_DATA.Loc value.
The Nearest_Loc must have existed when the observation was made. That is, an observation's related SWERB_DEPARTS_DATA.Date must be between the Nearest_Loc's related SWERB_GWS.Start and Finish (inclusive).
The SWERB_LOC_DATA.SWId of the location observation that was analyzed.
This column is unique, cannot be changed, and must
not be NULL.
The SWERB_LOC_CONFIDENCES.Confidence score of the location observation.
This column may not be NULL.
The SWERB_GWS.Loc whose related coordinates (XYSource) were determined to be nearest to this SWId's SWERB_DATA.XYLoc.
This column may not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
The Babase database uses a procedure called interpolation to update MEMBERS whenever the CENSUS table, or the BIOGRAPH.Birth, or BIOGRAPH.Statdate columns are updated. Interpolation extrapolates the group membership of individuals into days for which there is no actual observation of the individuals' whereabouts. It “guesses” in which group an individual is primarily, physically, located, given knowledge of the individual's group membership (or lack thereof) at given points in time, and records the result in MEMBERS. Thus, MEMBERS always has a row recording group membership for every day of every individual's life.
This section is comprised of 3 sub-sections. The first section introduces interpolation incrementally. Rules are presented in an informal fashion and examples and exceptions progressively developed. The second section is a formal specification of interpolation. The third section supplements the formal specification with expectations regarding the use of interpolation and brief descriptions of interpolation's implications. Most of the third section is a restatement of material already presented in the first section.
It is primarily by the field census records that Babase tracks group membership. However, despite its name, within the Babase database the the CENSUS table is the source of all group membership information and so contains data from sources other than just the field census records. Babase places rows in the CENSUS table to indicate presence in a group whenever any demography information is stored other tables.[180][181] Throughout this section it is to be understood that any sort of demographic information that results in CENSUS data are implied when the term census, or its plural, is used. Unfortunately, the term census is further overloaded. It is occasionally used in the colloquial sense, meaning present -- found when a group census was taken, the alternative being absent. It is hoped the meaning will be clear from context.
It is important to remember that censuses record absence from a group as well as presence in a group, that there are two mutually exclusive classes of CENSUS rows: absences, records of absence from specific groups on specific days; and “locating censuses”, records that place the individual in specific groups on specific days.
The premise of interpolation is that an individual is assumed to be in the group where observed for a period of 14 days to either side of the observation unless there's indication otherwise. To this end, interpolation keeps an individual in the group where a census locates him for a time period that is the shorter of:
Half of the time interval between the individual's next (or prior) census that finds the individual in any group.
Half of the time interval between the next (or prior) recorded absence from the group in which the individual was censused. Absences from other groups are ignored.
The 14 day Interpolation Limit. Given no other information, an individual is considered to remain (or have been) in the group where observed for 14 days following (or preceding) the date of observation.
Should the above process not place an individual in a group, the individual is placed in the unknown group; so long as the individual is alive on the day in question.
There are some subtleties to these rules, and there is further elaboration necessary to allow for “old style” CENSUS rows, which do not directly correspond with actual census taking, and other factors. But these rules are the foundation and we begin with them.
Interpolation is best described with the help of diagrams as it is all about computing and comparing time intervals of various lengths, which are easily represented in a diagram by lines of various lengths. We begin with the simplest case, censusing a single individual either present or absent in a single group. This simple case is elaborated on extensively to illustrate a variety of special cases such as birth, death, prolonged periods without observation, and so forth, before introducing the complexities of multiple groups into the example.
Tip
As the examples throughout this section are developed be sure to pay close attention to the diagrams' keys. At times the meaning of a symbol changes from diagram to diagram to reflect a subtlety.
Figure 4.1 shows a record
of one individual's censuses. The group, for the moment
we'll assume group 1, is censused 4 times
over a period of 11 days. One day the individual is
absent.
Figure 4.1. An Individual is Censused Present and Absent
One individual's census records
CENSUS: C C A C
Date: 1 2 3 4 5 6 7 8 9 10 11
Key:
C Censused present in group (group 1)
A Censused absent in group (group 1)
The first step in interpolation is to construct the various intervals from the given CENSUS rows. Figure 4.2 shows how interpolation “splits the difference” between presences and absences to construct two intervals for each locating census, one preceding the census and one following it. As the diagrams given here can only show a window in time and omit what falls outside that window, only one interval each is shown for the censuses taken on day 1 and day 11.
Figure 4.2. Interpolating From Presences and Absences
Interpolation intervals within a group
CENSUS: C C A C
Intervals: X---|---X---------| O |-----X
Date: 1 2 3 4 5 6 7 8 9 10 11
Key:
C Censused present in group (group 1)
A Censused absent in group (group 1)
X Known present in group (group 1)
O Known absent in group (group 1)
- Presumed in group (group 1)
| Midpoint between census takings
Interpolation creates MEMBERS rows that place the
individual in a group each day. Figure 4.3 shows how group membership
assignment is based upon the computed intervals. Because of
the absence, the individual is placed in group
9, the unknown group, on some
days.
Figure 4.3. Interpolating Group Membership
Intervals determine group membership
CENSUS: C C A C
Intervals: X---|---X---------| O |-----X
MEMBERS.
Group: 1 1 1 1 1 9 9 9 9 1 1
Origin: C I C I I I I I I I C
Date: 1 2 3 4 5 6 7 8 9 10 11
Key:
C Censused present in group (group 1)
A Censused absent in group (group 1)
X Known present in group (group 1)
O Known absent in group (group 1)
- Presumed in group (group 1)
| Midpoint between census takings
Figure 4.3 also introduces
the MEMBERS' Origin column. As can be
seen, the Origin column mimics the
corresponding CENSUS Status column on those days when
interpolation is not guessing group membership. Origin is
I on those day when
interpolation is guessing.
The MEMBERS' Interp column represents number of says from a census in which an individual was recorded as present in some known group. Interp is zero on those days when a census has located the individual. The recorded absence is reflected in the group, but is immaterial to Interp. Even though there's an absence, the Interp count is over the interval between the two locating censuses. Interp gets its value from a “split the difference” between censuses that record presence in the group, a different sort of “split the difference” than is used to determine into which group an individual should be placed. Figure 4.4 extends Figure 4.3, showing the computation of Interp. With this addition the interpolation has finished, the MEMBERS table can be constructed from the given CENSUS rows.
Figure 4.4. Computing Interp Values
The resulting MEMBERS rows
CENSUS: C C A C
Intervals
For Group: X---|---X---------| O |-----X
For Interp: X~~~|~~~X~~~~~~~~~~~~~~~|~~~~~~~~~~~~~~~X
MEMBERS.
Group: 1 1 1 1 1 9 9 9 9 1 1
Interp: 0 1 0 1 2 3 4 3 2 1 0
Origin: C I C I I I I I I I C
Date: 1 2 3 4 5 6 7 8 9 10 11
Key:
C Censused present in group (group 1)
A Censused absent in group (group 1)
X Known present in group (group 1)
O Known absent in group (group 1)
- Presumed in group (group 1)
~ Inside of interval
| Midpoint of interval
So far we have only explored the first 2 of the 3 fundamental interpolation intervals, those dealing with being censused present and absent. Before we elaborate further and examine the more complicated interactions between presences and absences let us dispense with the 14 day interpolation limit.
Figure 4.5 shows the effect of the
14 day interpolation limit. To save
space in this document, some days are removed from the
interval. There are no censuses, present or absent, on the
days omitted. As the “Date:” line shows, a
total of 33 days are examined, an entire month 31 days in
length and the first two days of the following month.
Again, we assume the censuses are taken in group
1.
Figure 4.5. The 14 Day Interpolation Limit
The shorter intervals are chosen
CENSUS: C C
C C Interval: X----- ... -----------|------- ... ---------X
14 Day Limit: X----- ... -------| |--- ... ---------X
MEMBERS.
Group: 1 1 ... 1 1 9 9 1 ... 1 1 1
Interp: 0 1 ... 13 14 15 15 14 ... 2 1 0
Origin: C I ... I I I I I ... I I C
Date: 1 2 ... 14 15 16 17 18 ... 31 1 2
Key:
C Censused present in group (group 1)
X Known present in group (group 1)
- Inside of interval
| Interval endpoint
Because the 16th and 17th are more than
14 days away from either census the
individual is placed in the unknown group on those
days. Days that are closer to the actual censuses are
interpolated into group 1. So, as the
rules require, the individual is interpolated into the
censused group for the shorter of the two time periods.
As before, all the interpolated MEMBERS
rows, those which do not correspond to an actual census,
have an Origin of
I. And as before, the
Interp column counts up from and
down to the actual censuses.
There are some exceptions to the rules as stated so far. Not surprisingly, interpolation will not presume to put an individual in a group, create a MEMBERS row, before the individual's Birth date.
The birth date is an exception in another fashion, it
locates the individual in his Matgrp like a special sort of census.
The rationale for this is that although the birth may not be
observed, the individual most certainly enters the group
when born. Further, this rule ensures that we have a row in
MEMBERS for every day the individual is
alive. When there is a regular census on the birth
date[182] the resultant MEMBERS row,
having a date matching the individual's birth date, is no
different from the individual's other MEMBERS rows that have dates which match the
individual's other census dates; they all have an Origin of C
and an Interp of
0. When there is no locating census on the birth date the resulting MEMBERS row still have a 0
Interp value, but have a Origin of
I, not
C. The Origin reflects the fact that there was
no actual census, while the Interp
shows that the individual was located that day. Figure 4.6 shows an individual that was not
censused on his birth date.
Figure 4.6. Interpolation at Birth
Individual born into group 1
CENSUS: B C C C
Intervals: X-----|-----X-|-X-----|-----X
MEMBERS.
Group: 1 1 1 1 1 1 1 1
Interp: 0 1 1 0 0 1 1 0
Origin: I I I C C I I C
Date: 1 2 3 4 5 6 7 8 9 10
Key:
B Born (into group 1)
C Censused present in group (group 1)
X Known present in group (group 1)
O Known absent in group (group 1)
- Presumed in group (group 1)
| Midpoint between census takings
Clearly, there are no MEMBERS rows
before the birth date, the individual is in his Matgrp on the day of his birth,
and the Interp value counts up from the birth date and
then down to the next census as though there were a
census on the birth date.
An individual is placed in his Matgrp on his birth date even when a regular census has an absence recorded for the individual on the date of birth.[183]
Another exception to the rules, or rather two exceptions, occur at the Statdate. You might expect that interpolation would not place a row after the individual's Statdate, and this is indeed true, but true only when the individual is dead. When an individual is alive, interpolation will place a row after the individual's Statdate, but only when there is a subsequent absence from the same group as the group in which the individual was censused.[184][185] While at first this may seem odd, the reasoning behind this behavior is clear -- the Statdate is not the last date on which there are data for the individual. This is elaborated below.
All the same, at times there is a reason to have interpolation halt at the Statdate. When individuals are alive the system should not try to interpolate into time periods for which data have yet to be entered, else-wise there would always be spurious interpolated MEMBERS rows which vanish as soon as additional data are entered. The trouble with creating such rows is that, although the interpolation is corrected and the rows disappear once data entry resumes, the use of these rows in analysis is always inappropriate. Such rows will exist at the end of every period of data entry, as there will always be a large number of living individuals found in their groups on the last census entered. The solution is to not create the rows.[186] When a living individual has no later absences from the group where last located, no absences from the group of his last locating census that post-date his last locating census, this is taken to mean that there are additional as yet unentered data on the individual. In this case interpolation stops on the day the individual was last found in a group. This situation is shown in Figure 4.7, where the last census taken found the individual in group 1 on day 5, and so this day is the individual's Statdate as well. There is no interpolation past the last census.
Figure 4.7. Alive and Present When Last Censused
Living individual with Statdate of 5
CENSUS: C A C
Intervals: X-----| O |-X
MEMBERS.
Group: 1 1 9 9 1
Interp: 0 1 2 1 0
Origin: C I I I C
Date: 1 2 3 4 5 6 7 8 9 10 11
Key:
C Censused present in group (group 1)
A Censused absent in group (group 1)
X Known present in group (group 1)
O Known absent in group (group 1)
- Presumed in group (group 1)
| Midpoint between census takings
In Figure 4.8 more data have been entered, the individual has been missing since the last census shown in Figure 4.7 above. As there have been no further censuses during which the individual was found the individual's Statdate is still day 5, although there is now subsequent interpolation. Notice that there are no MEMBERS rows created after day 7. When interpolating a living individual, after the Statdate there is no default placement of the individual into the unknown group.[187]
Figure 4.8. Alive and Absent in Last Census[188]
Living individual with Statdate of 5
CENSUS: C A C A A
Intervals: X-----| O |-X---------| O
MEMBERS.
Group: 1 1 9 9 1 1 1
Interp: 0 1 2 1 0 1 2
Origin: C I I I C I I
Date: 1 2 3 4 5 6 7 8 9 10 11
Key:
C Censused present in group (group 1)
A Censused absent in group (group 1)
X Known present in group (group 1)
O Known absent in group (group 1)
- Presumed in group (group 1)
| Midpoint between census takings
Although the only change between Figure 4.7 and Figure 4.8 is the entry into CENSUS of rows recording absence, that is enough
to signal that interpolation can go forward without creating
spurious MEMBERS rows -- rows likely
erased upon the entry of more data. It is important that
interpolation does go forward in this case, past the Statdate, as otherwise bias would be
introduced. The last C CENSUS would be interpolated differently from
all the other censuses. To be sure, there is bias
introduced in Figure 4.7 when
interpolation is cut short. But censoring bias at the end
of data collection is unavoidable, whereas we can avoid
introducing bias here.
Warning
So long as an individual is alive the last CENSUS to locate the individual ought be followed by a record of absence, an absence from the group where the individual was last found. To do otherwise, as must occur when there is simply no further data to be entered, is to introduce a bias into MEMBERS.
In Figure 4.9 there is no additional census information, but the individual's Status has been adjusted to mark the individual dead. A new Statdate value indicates the individual died on day 9 and interpolation is now up to and including the day of death. As is usual, when an individual's group membership cannot be determined he is placed in the unknown group.
Figure 4.9. Interpolation to Statdate When Dead
Dead individual with Statdate of 9
CENSUS: C A C A A
Intervals: X-----| O |-X---------| O
MEMBERS.
Group: 1 1 9 9 1 1 1 9 9
Interp: 0 1 2 1 0 1 2 3 4
Origin: C I I I C I I I I
Date: 1 2 3 4 5 6 7 8 9 10 11
Key:
C Censused present in group (group 1)
A Censused absent in group (group 1)
X Known present in group (group 1)
O Known absent in group (group 1)
- Presumed in group (group 1)
| Midpoint between census takings
Although Figure 4.9 does not show this, the 14 day interpolation limit applies when the individual is dead. When there are no absences after the last census and there are more than 14 days between the last census and the Statdate the individual is placed in the unknown group from the 15th day through the day of death.
The alert reader may have noticed that the above examples are carefully crafted so that the midpoint between presences and absences always falls between two days. What happens when there is an odd number of days in the interval so that the midpoint is a day exactly in between the endpoints, as occurs 3 times in Figure 4.10?
Figure 4.10. Midpoint Days
Intervals with an odd number of days
CENSUS: C A C C A C
Intervals: X---| O |-------X-|-X---| O |-X
Date: 1 2 3 4 5 6 7 8 9 10 11
Key:
C Censused present in group (group 1)
A Censused absent in group (group 1)
X Known present in group (group 1)
O Known absent in group (group 1)
- Presumed in group (group 1)
| Midpoint between census takings
The MEMBERS table has a 1 day precision, there is no way to be in a group in the morning and out of it in the afternoon, so on any one midpoint day the individual must either be in the group or out of it. Should the individual be in the group on midpoint day or out of it? The question is resolved using a property of the date itself. Briefly, the Julian dating system is a method of assigning every day a unique number. As a midpoint day is no more likely to be on one day than another, we can avoid bias by using whether or not the midpoint day falls on an even or an odd Julian date to resolve the problem.
Whenever interpolation is called upon to halve an interval between two CENSUS rows that contains an odd number of days then the “midpoint day” is assigned to the left, earlier, half of the interval when the Julian date of the midpoint day is even. A midpoint day is assigned to the right, later, half of the interval when the Julian date of the midpoint day is odd.
So, The Midpoint Rule resolves the issue by adjusting the intervals as shown in Figure 4.11. The intervals are no longer perfectly halved. On the midpoint day there is no preference either for or against interpolating the individual into the group censused.
Figure 4.11. The Midpoint Rule Adjusts Intervals
Intervals with an odd number of days
CENSUS: C A C C A C
Intervals: X-----| O |---------X-|-X-| O |-X
MEMBERS.
Group: 1 1 9 9 1 1 1 1 9 9 1
Interp: 0 1 2 3 2 1 0 0 1 1 0
Origin: C I I I I I C C I I C
Julian Date: 1 2 3 4 5 6 7 8 9 10 11
Key:
C Censused present in group (group 1)
A Censused absent in group (group 1)
X Known present in group (group 1)
O Known absent in group (group 1)
- Presumed in group (group 1)
| Interval endpoint
Having dispensed with the various elaborations and exceptions that occur in unusual cases it is time to return to the fundamentals of interpolation and examine what happens when an individual moves between groups. What comes into play are the first 2 of the 3 interpolation intervals. Recall:
Interpolation keeps an individual in the group where a census locates him for a time period that is the shorter of:
Half of the time interval between the individual's next (or prior) census which finds the individual in any group.
Half of the time interval between the next (or prior) recorded absence from the group in which the individual was censused. Absences from other groups are ignored.
Figure 4.12 shows a record of one individual's censuses. He, a male, is censused in 2 groups, group 1 and group 2. The census records for each group reflect both presence in the group and absence from the group.
Figure 4.12. An Individual is Censused in 2 Groups
One individual's census records
Group 1: C C A C A
Group 2: A C C
Date: 1 2 3 4 5 6 7 8 9 10
Key:
C Censused present
A Censused absent
Figure 4.13 shows what would happen if interpolation worked with each group separately. There are conflicts, days when the individual is in both groups. Something else must be done.
Caution
Figure 4.13 is an example of an interpolation method that does not work. The method shown in the figure is not one Babase uses when interpolating.
Figure 4.13. Interpolating Each Group Separately
One individual's census records
Group 1: C C A C A
Group 2: A C C
Group 1 Interpolating just group 1
CENSUS: C C A C A
Intervals: X---|---X---------| O |-X-| O
Group 2 Interpolating just group 2
CENSUS: A C C
Intervals: O |---------X-------|-------X
Date: 1 2 3 4 5 6 7 8 9 10
Key:
C Censused present
A Censused absent
X Known present
O Known absent
- Presumed present
| Interval endpoint
The solution is return to the interpolation fundamentals. We begin by taking a closer look at the way we have been diagramming intervals. In Figure 4.13 the first group has 3 locating census and 2 absences, and yet we've diagrammed the resultant intervals on a single line. The interpolation fundamentals tell us to obtain 2 pairs of intervals for each locating census. A “halfway to census” pair of intervals and a “halfway to absence” pair of intervals. Figure 4.14 takes the CENSUS rows of the first group shown in Figures 4.12 and 4.13 and does this for each locating census. In Figure 4.14 the CENSUS rows of days 1, 3 and 9 each have their own sections detailing the intervals to the nearest censuses and intervals to the nearest absences. The lines labeled Presence show the intervals that are halfway from each locating census to the next. The lines labeled Absence show the intervals that are halfway from each census to the nearest absence. This detailed breakdown is followed by a composite interval diagram of the familiar type encountered in figures 4.2 through 4.13 above. It should be clear that we have arrived at the “composite” form of the interval diagram by following the fundamentals, the composite is made up of the shorter of each census's intervals. The result is correct, the composite constructed in Figure 4.14 is identical to the one shown previously in Figure 4.13. It had better be, or else the interpolations of Figure 4.13 would be in conflict with the fundamental interpolation rules.
Figure 4.14. A Closer Look at Intervals
CENSUS rows from group 1
CENSUS: C C A C A
Day 1 Intervals by presence and absence
Presence: X---| X
Absence: X-------------| O
Day 3 Intervals by presence and absence
Presence: X |---X-----------| X
Absence: X---------| O
Day 9 Intervals by presence and absence
Presence: X |-----------X
Absence: O |-X-| O
Combining the shorter intervals
Interval: X---|---X---------| O |-X-|
Date: 1 2 3 4 5 6 7 8 9 10
Key:
C Censused present
A Censused absent
X Known present in same group
x Known present in different group
O Known absent in same group
- Inside of interval
| Interval endpoint
The intervals in Figure 4.14 did not have to be grouped by censused day, they could have been grouped by Presence and Absence or any other way. For each set of locating censuses we can always split out the “halfway to census” intervals from the “halfway to absence” intervals, group them any way we like, and later use the interpolation fundamentals to recombine them, without affecting the result. This has not been necessary so far, but it is essential if we are to correctly interpolate when an individual moves between groups, as above in Figure 4.12: “An Individual is Censused in 2 Groups”. We must return to the fundamentals to make sense of interpolation. Rather than trying to combine the results of interpolating the groups separately, as was done in Figure 4.13: “Interpolating Each Group Separately”, instead combine the results of interpolating the presences in all the groups with separate interpolations of the absences in each group. Each time a census finds an individual in a group, separately compute both the interval halfway to the nearest census that finds the individual in any group and the interval halfway to the nearest absence from the particular group being censused.[189]In Figure 4.15, this method is applied to the data first seen in Figure 4.12. For clarity the intervals surrounding the censuses that belong to one group are shown separately from those belonging to the other group.[190] The lines labeled Presence show the intervals that are halfway from each census to the nearest census that finds the individual in any group. The lines labeled Absence show the intervals that are halfway from each census to the nearest absence in the same group. Censuses with no neighboring absence do not have this latter sort of interval shown.[191]
Figure 4.15. Presence and Absence Interpolated Separately
One individual's census records
Group 1: C C A C A
Group 2: A C C
Group 1 The intervals of group 1's censuses
Presence: X---|---X-----| x |-----X-| x
Absence: X---------| O |-X-| O
Group 2 The intervals of group 2's censuses
Presence: x x |-----X-----| x |-X
Absence: O |---------X
Date: 1 2 3 4 5 6 7 8 9 10
Key:
C Censused present
A Censused absent
X Known present in same group
x Known present in different group
O Known absent in same group
- Inside of interval
| Interval endpoint
Figure 4.16 shows how interpolation combines the “presence” and “absence” intervals by choosing the shorter of the two to as the period during which the individual is assumed to be in the group where censused. The line labeled Used contains the shorter of each census's two intervals.[192]
Figure 4.16. Combining Presence and Absence Intervals
One individual's census records
Group 1: C C A C A
Group 2: A C C
Group 1 The intervals of group 1's censuses
Presence: X---|---X-----| x |-----X-| x
Absence: X---------| O |-X-| O
Used: X---|---X-----| |-X-|
In Group: 1 1 1 1 ? ? ? ? 1 ?
Group 2 The intervals of group 2's censuses
Presence: x x |-----X-----| x |-X
Absence: O |---------X
Used: |-----X-----| |-X
In Group: ? ? ? ? 2 2 2 ? ? 2
Date: 1 2 3 4 5 6 7 8 9 10
Key:
C Censused present
A Censused absent
X Known present in same group
x Known present in different group
O Known absent in same group
- Inside of interval
| Interval endpoint
Having interpolated the intervals surrounding each census, determining the final group membership is a straightforward matter of placing the individual in the unknown group when there's no where else to put him. Figure 4.17 shows this process. All that remains is to compute the Interp values in the usual fashion, by ignoring absences and counting distance from the nearest census. In Figure 4.17 the intervals between locating census are shown, labeled For Interp, to support the Interp values given.
Figure 4.17. Group Membership Given Multiple Groups
One individual's census records
Group 1: C C A C A
Group 2: A C C
Group 1 The intervals of group 1's censuses
Used: X---|---X-----| |-X-|
In Group: 1 1 1 1 ? ? ? ? 1 ?
Group 2 The intervals of group 2's censuses
Used: |-----X-----| |-X
In Group: ? ? ? ? 2 2 2 ? ? 2
Intervals between locating censuses
For Interp: X~~~|~~~X~~~~~|~~~~~X~~~~~|~~~~~X~|~X
MEMBERS.
Group: 1 1 1 1 2 2 2 9 1 2
Interp: 0 1 0 1 1 0 1 1 0 0
Origin: C I C I I C I I C C
Date: 1 2 3 4 5 6 7 8 9 10
Key:
C Censused present
A Censused absent
X Known present in same group
- Presumed present
~ Inside of interval
| Interval endpoint
By now it should be clear that interpolation[193] is a function over CENSUS row sets. It is a function, for every input you get exactly one output. It takes sets of CENSUS rows as input. Because sets are unordered you can put CENSUS rows into the database in any order and the result will be the same. And, because it is a function, you can re-interpolate the same CENSUS rows as many times as desired without altering the final result.
It should also be clear why interpolation always chooses to use “the shorter interval”, and why this always produces the “correct” result. The shorter interval is short for a reason, there is some reason to believe the individual is not in the group else-wise the interval would be longer. Further, every time the shorter interval is chosen a possible overlap with another interval from a different locating census is eliminated. By always choosing the shorter interval interpolation insures that the interpolation of any two locating censuses will not conflict.
In addition to that most important distinction which
classifies CENSUS rows into absent and
locating censuses
there is a second distinction which further divides locating
censuses into those which interpolate and those which do
not. Those CENSUS rows that record
observational data are interpolating censuses; those
with Status values of
C,
D and,
M.[194] (All of the previous examples have concerned
CENSUS rows of this type.) The remaining CENSUS.Status values
indicate that the CENSUS row is the result
of analysis, all of the “old style”, that is
“historical”, CENSUS.Status values and the
N manual Status
value. These are the
non-interpolating censuses.
This further division of locating censuses into
interpolating and non-interpolating, the division between
raw and already analyzed data, leads to the final refinement
to the interpolation procedure. We do not want
interpolation to produce re-analyzed results from already
analyzed data. Interpolation occurs only between
“regular”, that is to say interpolating,
censuses (and to the birth date as a special case).
“Non-interpolating” census rows are copied
directly from CENSUS to MEMBERS, CENSUS.Status becomes MEMBERS.Origin, and
Interp is set to NULL. When a
non-interpolating census is found on the birth date, the
birth date will not interpolate.
Interpolation looks at “regular” census rows and attempts to guess the individual's location on those days when there are no observations. It does so by looking at the intervals between the “regular” censuses. Finding non-interpolating CENSUS rows, that is to say already analyzed data, on one of these intervals breaks the assumptions interpolation uses in its “guessing”. The previously analyzed data point could be there for any reason at all, and there's no point in pretending it's not there either. What interpolation does is give up. It interpolates up to the offending data point and then stops.[195] After that it still creates rows in MEMBERS, but it does not attempt to make guesses about where to place an individual or what the interpolated row means.
Note
This situation is not expected to occur, or, rather, whenever there are non-interpolating CENSUS rows between interpolating censuses, the non-interpolating CENSUS rows are expected to be contiguous over the entire interval between the interpolating censuses. So, the expected cases are the trivial degenerate ones. None the less, such situations probably do occur in the existent data. It would probably best to either require the expected behavior, or to get rid of all the pre-analyzed CENSUS rows and replace them with raw data. Especially given the design problems pointed out below.
Regardless, non-trivial examples are presented here so that a complete understanding of interpolation can be developed.
Figure 4.18 shows that the
3 fundamental interpolation intervals are shortened when a
non-interpolating census is found between interpolating
censuses. The intervals for each locating census
are examined separately. The non-interpolating census has
no interpolation intervals. The intervals of the
interpolating censuses are truncated, reduced to the
interval between the interpolating and non-interpolating
censuses. By this means a portion of the diagram, days 4
and 5, are blocked from interpolating into the group. If
there were no N
census, the Absence interval would
be day 1's shortest interval, and days 4 and 5 (as well as
day 3) would interpolate into the group. (Notice that day
1's Absence interval has a midpoint
day, day 5, and that it would have been included in the
interval.) Interpolation is prevented from placing
individuals in the group of their interpolating census on
the “far side” of non-interpolating
censuses.
Figure 4.18. Pre-Analyzed Data Truncates Interpolation Intervals
CENSUS rows from group 1
CENSUS: C N A C
Day 1 Intervals per fundamental type
Presence: X-----| N X
Absence: X-----| N O
14 Day Lim: X-----| N
Day 3 Intervals per fundamental type
Presence: N
Absence: N
14 Day Lim: N
Day 12 Intervals per fundamental type
Presence: X N |---------------------X
Absence: N O |-----X
14 Day Lim: N |---------------------------------X
Julian Day: 1 2 3 4 5 6 7 8 9 10 11 12
Key:
C Censused present in group (group 1)
N Manual entry,
present in group but non-interpolating (group 1)
A Censused absent in group (group 1)
X Known present in group (group 1)
O Known absent in group (group 1)
- Inside of interval
| Interval endpoint
In Figure 4.19 the shortest intervals of each locating census have been chosen and combined; the result is the line labeled For Group. This is then used to determine group membership.
The interesting part of Figure 4.19
is the computation of the Interp
values. The “halfway to census” intervals of
Figure 4.18 have been combined
and labeled For Interp. Recall
that it is these intervals that are used to compute the
Interp values. The
N census has created
a “gap” in interpolation, clearly shown on the
For Interp line as running from day
3 through day 6. Over this interval interpolation's
assumptions have been violated and it does not know what to
do. The group membership is easy. On day 3, the day of the
N census it can
simply copy the CENSUS row's Grp and Status
into the appropriate MEMBERS columns in
the same fashion it would for any other locating census. On days 4 through 6 it can do what it
usually does with group membership when it does not know
where to locate an individual, it places the individual in
the unknown group with a Origin
of I. On days 3 through
6 interpolation has no way of knowing how far away the day
is from the nearest locating census, which is what
is supposed to go in the Interp
column. Due to this lack of information it assigns the Interp column a value of NULL, no
data, on this interval.
Figure 4.19. Pre-Analyzed Data Interrupts Interpolation
An individual is censused
CENSUS: C N A C
Intervals
For Group: X-----| N O |-----X
For Interp: X~~~~~| |~~~~~~~~~~~~~~~~~~~~~X
MEMBERS.
Group: 1 1 1 9 9 9 9 9 9 9 1 1
Interp: 0 1 5 4 3 2 1 0
Origin: C I N I I I I I I I I C
Date: 1 2 3 4 5 6 7 8 9 10 11 12
Key:
C Censused present in group (group 1)
N Manual entry,
present in group but non-interpolating (group 1)
A Censused absent in group (group 1)
X Known present in group (group 1)
O Known absent in group (group 1)
- Presumed in group (group 1)
~ Inside of interval
| Interval endpoint
When looking at Figure 4.19, one way
to explain what happens to Interp
is to say that it is fixed at NULL over that portion of
the day 1 census's “halfway to census” interval
that was truncated because the
N row showed up.
(See Figure 4.18.) Effectively,
as MEMBERS Interp counts up with increasing
distance from the interpolating census, the count is fixed
at NULL upon encountering a non-interpolating census until
the point is reached at which counting back down to the next
interpolating census begins, at which point the count
downward resumes as though never interrupted.[197]
The approach interpolation
takes, in some sense, attempts to minimize the disturbance
created when already analyzed census data are mixed in with
raw census information. However, as can be seen in Figure 4.19, it is not entirely successful.
Although day 7, for example, has an Interp value indicating it is 5 days
away from a census, it is really 4 days away from the
N census. If the
N CENSUS does really represent a census, then day
7's Interp value is wrong. And
the problems are not restricted to Interp values. Is it really true that
days 4 and 5 should be assigned to the unknown group? If so
then why aren't there
N rows that say so?
Day 2 is even more disturbing. There is no diagram for
this, but suppose the
N census found the
individual in a different group. Figure 4.18 would be unchanged, all of
day 1's intervals would be truncated at the
N census. The effect
would be more clear if the interval between the preceding
C census and the following
N census were larger,
but consider that day 2, by the midpoint rule, would be
“assigned” to the
N census. That means
that if the N census
really does represent a census in a different group, that
day 2 should be assigned to that group, not to group
1.
Note that, in the general case, even though the
“halfway to census” interval does not determine
group membership (all the intervals are truncated, leaving a
“gap” in which interpolation defaults to the unknown group), whether this interval has a midpoint day,
and if so where it falls, does matter
to the computation of Interp. If
the midpoint day happens to fall into the side of the
interval containing the non-interpolating census then the
Interp value will be NULL.
Otherwise, it will have a value representing the number of
days to the nearest locating,
and interpolating, census.
Incorporating the above safety checks into the rules we already have, ensuring that data are not re-analyzed, produces the actual interpolation rules.
Using these rules interpolation creates rows in MEMBERS based on the information it finds in CENSUS, and the BIOGRAPH columns Birth, Matgrp, Statdate and Status.
CENSUS Rows Are Either Absences, Interpolating, or Non-Interpolating
Interpolation partitions all CENSUS rows into one of 3 categories:
CENSUS rows which indicate absence from a group.
Those CENSUS rows that record observational data are interpolating censuses; those with Status values of
C,Dand,M.The remaining CENSUS.Status values indicate the CENSUS row is the result of analysis. These rows, all of the “old style”, that is “historical”, CENSUS.Status values and the
Nmanual Status value, are not re-analyzed and so do not interpolate.
For convenience, the CENSUS rows that are not absences, the interpolating and the non-interpolating censuses, are termed “locating censuses”.
Censusing Assigns Group Membership
On those days when an individual is censused in a group, when there is a locating CENSUS row, a row is created in MEMBERS to place that individual in the group on the given day. The Origin value is the CENSUS row's Status value. When the CENSUS row is interpolating the Interp value is
0. When the CENSUS row is non-interpolating the Interp value isNULL.Interpolation places an individual in the group into which he is censused, the Grp of an interpolating CENSUS row (Status values
C,D, andM), on the days to either side of the census being interpolated for a time period that is the shorter of:The Halfway to Census Interval
Half of the time interval between the individual's next (or prior) locating and interpolating census, which may locate the individual in any group.
The Halfway to Absence Interval
Half of the time interval between the next (or prior) recorded absence, considering only absences from the same group in which the individual was censused. Absences from other groups are ignored.
The 14 day Interpolation Limit
Given no other information, an individual is considered to remain (or have been) in the group where observed for 14 days following (or preceding) the date of observation.
The resulting MEMBERS rows have an Origin of
Iand an Interp value of the number of days difference between the MEMBERS row's Date and the date of the nearest locating census; Interp values count up over the The Halfway to Census Interval as the distance from the interpolated census increases. An interpolated MEMBERS row falling on the day after a census has an Interp of 1, the day after that the Interp is 2, and so forth, assuming, of course, the individual has no other nearby CENSUS rows.This rule qualifies how interpolation assigns the halfway point between two CENSUS rows in The Halfway to Census Interval and The Halfway to Absence Intervals, above, when the number of days in the interval cannot be divided into equal halves. Whenever interpolation is called upon to halve an interval between two CENSUS rows that contains an odd number of days then the “midpoint day” is assigned to the left, earlier, half of the interval when the Julian date of the midpoint day is even. A midpoint day is assigned to the right, later, half of the interval when the Julian date of the midpoint day is odd.
This rule declares a live birth to be the equivalent of an interpolating census, one that indicates presence in the individual's Matgrp. fetal losses, individuals with
NULLSnames, are not considered births and are never interpolated. An individual is placed in his Matgrp on his birth date even when a regular census has an absence recorded for the individual on the date of birth. In this case interpolation always entirely ignores the absence and will not use such an absence to compute a Halfway To Absence Interval.When there is a locating census on the birth date, the MEMBERS row interpolation creates is like that made for any other locating census with the given Status. But, when there is no locating census on the birth date the resulting MEMBERS row has a Origin of
I(and an Interp of0as any census with a Status ofCwould have.) Aside from theirIOrigin value, births interpolate as would any CENSUS with aCStatus.No Data Implies Unknown Group Membership
On days when none of the above rules serve to place an individual in a group, the individual is placed in the unknown group. The resulting MEMBERS rows have an Origin of
Iand an Interp value of the number of days difference between the MEMBERS row's Date and the date of individual's nearest interpolating census.[198]Interpolation will not place a row in MEMBERS before an individual's Birth date.
When an individual is dead, interpolation will not place a row after the individual's Statdate.
Data Entry Cessation Stops Interpolation of Living Individuals.
When an individual is alive, interpolation will create rows after the individual's last locating census only when there are subsequent absences; absences, that is, from the group in which the individual was censused.[199] In this case, unlike above, no data does not imply unknown group membership; such rows are created only so long as the individual is interpolated into the group of his last locating census. When a living individual has no absences after their last locating census, absences from the group of their last locating census, interpolation assumes that there is further data available which has yet to be entered and interpolation stops at the last locating census.
Interpolation is only done to regular, that is interpolating, CENSUS rows; data that were collected in the field. Other data, the “non-interpolating” census rows that represent the result of prior analysis, do not interpolate; they are copied directly from CENSUS to MEMBERS, CENSUS.Status becomes MEMBERS.Origin and Interp is set to
0. Further, when a non-interpolating census is found on one of The 3 Interpolation Intervals the interval is shortened enough that the non-interpolating census is no longer on the interval. When a non-interpolating census is found on a birth date, the birth date does not interpolate.The MEMBERS Interp column is fixed at
NULLon the interval from the non-interpolating census row through the “midpoint” end of The Halfway to Census Interval, endpoints included.[200] Here we are speaking of The Halfway to Census Interval as computed, not a Halfway to Census Interval shortened in the preceding paragraph.
It is expected that all non-interpolating CENSUS rows, that is to say CENSUS rows produced
by prior analysis, will be clustered in contiguous intervals
with “regular” census rows at the endpoints.
This is particularly expected of “old style”
census rows from before Babase, as they precede all
“regular” census data, but is also expected of
the N
non-interpolating, manual, Status
code, should it ever be used. If these expectations are born
out, the Data are not Re-Analyzed rule will
never be invoked.
There are some not-quite-obvious implications given these interpolation rules:
The only rows in MEMBERS that have an Origin of
I, and an Interp of0, and are not placed in the unknown group are birth dates. Not every birth date will have an associated MEMBERS row with these values, as some birth dates have locating censuses, but MEMBERS rows with these values will be birth dates.Living individuals, but not dead ones, can have MEMBERS rows created by the interpolation procedure that locate the individual in a group on a date later than the individual's Statdate.[201]
So long as an individual is alive the last CENSUS to locate the individual ought be followed by a record of absence, an absence from the group where the individual was last found. To do otherwise, as must occur when there is simply no further data to be entered, is to introduce a bias into MEMBERS.
Aside from births, the only other rows in MEMBERS with an Origin of
Iand an Interp of0are those in the unknown group which were created by the “Data are not Re-Analyzed” rule above.As fetal losses, individuals with
NULLSnames, cannot appear in CENSUS, are not considered a live birth, and always have their birth date equal to their Statdate, they never have MEMBERS rows associated with them.When computing Interp values from The Halfway to Census Interval The Midpoint Rule is usually immaterial. However, when non-interpolating censuses affect the interpolation The Midpoint Rule can be the factor that determines whether or not a MEMBERS row has a
0Interp value or not.
Sometimes, an individual’s group membership in MEMBERS shows them temporarily disappearing from a group. This might mean that the individual is preparing to leave the group, but there are alternative explanations that do not involve such a dispersal. Even when physically absent from a group, an individual may remain socially present there. After the system uses interpolation to estimate an individual's physical presence each day of their life (each row in MEMBERS), the system may analyze those data to estimate the individual’s social presence, or residency, on each of those days. These residency analyses "smooth out” temporary absences and provide an objective method for identifying when an individual has dispersed from a group.
Note
In this context, “disperse” is used differently than it is in the DISPERSEDATES table, where a “dispersal” is strictly defined as the date when a male leaves his maternal group. In this section, a “dispersal” is any case where an individual of any sex leaves a known group for a significant period of time.
Caution
Unlike interpolation, residency (and supergroup) information is not automatically updated by the system. The data managers are expected to run one or more of the data analysis procedures to update residency information whenever changes to Babase data might affect residency.
In general, an individual becomes resident in a group on a particular date if interpolation places them in the group more than it places them in any other group in a period beginning that date and extending through the next 28 days. They will remain resident in the group until the end of the last consecutive 29-day (“today” + 28 days) window that meets this criterion. The 29-day interval was chosen because interpolation can place an individual in a group for up to 14 days on either side of the date on which the individual was censused present in the group. 29 days is the longest period during which an individual can be interpolated into a group with a single census.
The residency rules are asymmetric in that there is one set of rules for establishing residency and another for maintaining residency. It is not always possible to determine if an individual is resident on any given day without knowing about the determination of the individual's residency on surrounding days.
Some specifics of the group residency rules differ
depending on the density of available census data for the
individual. When the MEMBERS.LowFrequency is TRUE, the process used to
determine an individual’s residency for that day may be based on
a slightly different set of rules. Briefly: if an individual is
a resident of a group, is not seen for an extended period, and
is still in that group the next time both the individual and the
group are seen, then the individual is presumed to have remained
resident in the group throughout the time that they were not
seen.
Residency is never assigned in groups 9.0 ("Unknown") and 10.0 ("Alone"), as these two GROUPS.Gid values explicitly do not represent actual social groups.
For brevity in this chapter, group identity is discussed as though it is absolute: a social group of individuals has a particular name or identity, and that group’s identity never changes. That presumption is often incorrect. Groups may gradually divide into subgroups, and multiple smaller groups may gradually assemble into a single larger group. This section describes how the residency rules determine the group in which an individual is assigned residency, especially during those periods of group fission or fusion when an individual’s presence in one group might be evidence for residency in another.
During fission and fusion periods, residency is determined with respect to the parent group(s). After fission/fusion completes — on the GROUPS.Permanent date of the daughter group(s) — individuals become resident in the daughter group(s). Consequently, all residency rules are with respect to the parent group(s) during periods of fission or fusion and otherwise are with respect to the current group[202]. There is one exception.
Shortly after a fission ends, some individuals may continue to float between the daughter groups. While the fission is still recent, these visits should still be treated as being in the same group. For the first 28 days after the fission ends, being in any daughter group is evidence for residency in both[203]. Still, during this period the system can only assign residency in one of the daughter groups. The group of residency is whichever group is visited first in this period.
To implement these rules in a way that is agnostic to the presence/absence of a group fission or fusion, the system uses the MEMBERS.Supergroup and Delayed_Supergroup columns — never the Grp — when determining presence in a group. On two given dates X and Y (where X < Y but X + 28 > Y), an individual is considered to be in the same group if any of the following conditions are met:
| If... | Then... |
|---|---|
| Supergroup on X = Supergroup on Y | The Grp on these two dates are identical, or they're two non-Permanent subgroups of the same group and this is during their fission |
| Supergroup on X = Delayed_Supergroup on Y | One of the above, or X is in a parent group during a fission or fusion and Y is in a daughter group after the fission/fusion |
| Delayed_Supergroup on X = Delayed_Supergroup on Y | One of the above, or both dates are ≤ 28 days after a fission and each of these dates are in different daughter groups of that fission. |
Note
This system is not especially robust to individuals switching between parent groups during a fusion. Special consideration is needed when adding those data to CENSUS, as discussed above.
When an individual is determined to be resident on a particular date and the individual is present in that group, the MEMBERS.GrpOfResidency is the individual’s Supergroup for that Date. If the individual was not present in the group on that date, the GrpOfResidency is the Supergroup of the first subsequent row in which the individual is present in the group.
Example 4.4. Determining the GrpOfResidency when absent
An individual is resident in Hook’s group, which is in the process of dividing into two new groups: Linda's and Weaver's. One week before the fission ends, the individual is present in (his MEMBERS.Grp is) Hook's on Monday, Linda's on Tuesday, unknown on Wednesday, and Weaver's on Thursday. Because this is before the fission ended, residency can only be obtained in the parent group. Therefore, the Supergroup (and the GrpOfResidency) on Monday, Tuesday, and Thursday is Hook's. On Wednesday, the supergroup is unknown, so the system looks to the Supergroup on the first subsequent day in Hook's/Linda's/Weaver's to get the GrpOfResidency. In this case, that would be Thursday, in Weaver's. The Supergroup on Thursday is Hook's, so the GrpOfResidency for Wednesday is Hook's.
A special case occurs when an individual retains residency on a date that they are not present in the group, but the Supergroup of the next “present” row is a group that was not yet permanent on the date of the individual’s absence. This happens at the end of fissions and fusions when the “not present” day is before the parent group(s) ceased to exist, and the next “present” day is after the daughter group(s) became Permanent. In this situation, the system uses the Delayed_Supergroup — not the Supergroup — of the first subsequent MEMBERS row in which the individual is present in the group.
Example 4.5. Resident in a nonexistent group
The same individual from the previous example is still in Hook’s group, the fission of which is just about to end. On Friday, the last day of the fission — when he's still a resident in Hook's group — he is absent. On Saturday — the next day that he’s present — he will be in Linda’s group, which at that point will be Permanent. Usually, when determining the GrpOfResidency for an absent day like Friday, the system would look at the Supergroup for that next "present" day ("Linda's") and use it as Friday's GrpOfResidency. But Linda's group wasn't Permanent on Friday, so the individual can't be resident there that day. Instead, the system uses Saturday's Delayed_Supergroup: Hook’s.
In another special case, an individual is not present in the group on a date and is not seen in that group again, but retains residency there because 1) the date is on or shortly before the individual's Statdate and 2) the individual's Status is a Residency_Special_Case. (See Statdate is (also) special for more details.) When this occurs, the system cannot look forward to the next "present" date and must instead look back. The GrpOfResidency is that of the most recent date that the individual was present in the group. In the rare case that the GrpOfResidency ceases to exist during this "special case" period, the new GrpOfResidency is whichever daughter group was most recently visited (in the individual's MEMBERS.Grp). If no such group is found, the new GrpOfResidency will be whichever daughter group is numerically first[204]. It is an error if the system still cannot determine a GrpOfResidency[205].
In principal, an individual is resident in a group for a given period of time if they are present in that group more than they are absent from it. To assess an individual’s presence/absence, the system often uses what will hereafter be referred to as the “15/29 test”.
For a given date, the system investigates the 29-day “window” that begins on that date and ends after the subsequent 28 days. If the individual is present in a group for at least 15 of those 29 days, the individual has “passed” the test. The individual will likely — but not necessarily — be assigned residency in the group. How the system uses this information and whether the individual actually is a resident on the given date are context-dependent, explained later.
While performing this test, the system also counts the
number of distinct dates in the window in which the individual
was censused in any group, including both
“present” and “absent” censuses. When this number is
3 or fewer, the pertinent MEMBERS.LowFrequency is
TRUE. Otherwise, it’s FALSE.
When there are fewer than 28
days after the given date in MEMBERS, the
individual must still be present for at least
15 of those days to pass the test, and there
must still be > 3 census dates to
have a LowFrequency of
FALSE.
To be clear: the 15/29 test alone does not determine whether or not an individual is resident in a group on a given date. The test is an important part of how residency is determined, but there are other factors that also affect that determination.
Residency for each individual is calculated via day-by-day iteration through the individual’s life history in MEMBERS. The system begins on the individual’s BIOGRAPH.Entrydate and continues chronologically onward through the individual's Statdate. While iterating through each date, the system tracks whether or not the individual is in an ongoing bout of residency and deploys a concomitant series of tests. When the individual is not in an ongoing bout, the system will try to obtain residency. When there is an ongoing bout, the system will try to retain residency.
When considering the 29-day window that begins on a specific date, the process of obtaining residency asks if the individual was resident in the group beginning on day 1. In contrast, the process of retaining residency asks if the individual was still resident on day 29. That is, obtaining residency is about finding the beginning of a residency bout, while retaining residency is about finding the end. These rules are discussed in greater depth below.
After the system finishes trying to obtain or retain residency for an individual on a date, the MEMBERS.Residency, LowFrequency, and GrpOfResidency for the row with that Date are populated.
When a continuous period of residency ends, the system also adds a row to the RESIDENCIES table for the entire period, or "bout". The Start_Date and Finish_Date are populated as expected. The Start_Status and Finish_Status are populated as discussed below.
As mentioned earlier, rows in MEMBERS
from days before the Entrydate or
after the Statdate are not analyzed
for residency. When a residency analysis is performed, the
Residency column for those rows is
set to X. Thus, it can safely be assumed
that when an individual has any MEMBERS rows
with a NULL Residency, that
individual's residency information is in need of an
update.
When the system iterates over a date on which the individual's residency has not yet been determined, the system will try to obtain residency for the individual on this date. If the individual is present in a real group — not 9.0 or 10.0 — on that date, the system performs a 15/29 test to determine if residency is obtained in that group. The putative end of the residency — the last day of the 29-day window in which the individual was present in the group, hereafter referred to as the putative last date — will likely be extended onward as the system iterates through subsequent dates and tries to retain residency.
While obtaining residency, it is not sufficient to merely be present in the group for 15 of the 29 days; the individual must also be present in the group on the first day of the 29-day window. Otherwise, individuals could become resident in a group up to 14 days before they are first present there.
If the individual passes the 15/29 test, their MEMBERS row for day 1 is updated to indicate that they’re resident: the Residency is set to R, the LowFrequency is populated according to the 15/29 test’s result, and the GrpOfResidency is that row’s Supergroup.
If the individual doesn’t pass the
15/29 test, or the individual is in group
10.0 on the date and didn’t attempt to obtain
residency there, the row is updated to indicate that they’re
not resident: the Residency is set
to N and the LowFrequency and GrpOfResidency are NULL.
If the individual is in group 9.0 on this
date and therefore can’t attempt to obtain residency, the
row’s Residency is set to
U and the LowFrequency and GrpOfResidency are NULL.
When residency is first obtained, the system categorizes how the residency period began. Later, when the residency ends, that information will be inserted into the RESIDENCIES.Start_Status column for the residency period. See RESIDENCIES.Start_Status for more information about the possible values.
In some cases, an individual's residency can be inferred from the way that the individual entered the study population. At birth, for example, an infant does not need to "earn" residency by being present for the requisite number of days; the infant simply is intuitively a resident of the group from the beginning. When attempting to obtain residency on the individual's Entrydate, the system allows the possibility that the decision of whether or not to obtain residency might depend on the individual's Entrytype.
When the individual's Entrytype is indicated in the ENTRYTYPES table as a Residency_Special_Case (when the
Residency_Special_Case is
TRUE), the individual automatically obtains residency in
the group in which they were present on the Entrydate. They do not need to pass
the 15/29 test; their residency in the
group is obtained solely because of their Entrytype. The only exception to
this: if the individual is in group 9.0 or
10.0 on their Entrydate, then they will not obtain
residency[206].
Individuals with any other Entrytype are not automatically assigned residency on their Entrydate. They may still become resident on this date, but they must pass the 15/29 test to obtain it, as usual.
When determining LowFrequency, the system has more
special treatment for individuals who were born into the
study population. To be clear: this is only for
individuals whose Entrytype is
B, not for all Entrytypes that are
Residency_Special_Cases. It
is presumed that if the group was being watched frequently
enough to know the individual's date of birth then it must
not have occurred during a low frequency period. Because
of this, in the MEMBERS rows for those
individuals' Entrydate and
subsequent 28 days, any days
where the individual is a resident and part of the bout
that began on the Entrydate
have their LowFrequency
automatically set to FALSE.
Note
These special provisions may seem unnecessary.
Most individuals whose Entrytype is a Residency_Special_Case would
likely obtain residency there anyway. For most
individuals whose Entrytype
is B, the 15/29 test
likely would have determined that the individual's LowFrequency was FALSE in those
early days anyway. Also, in rare occasions an
infrequently visited group might
happen to be visited on a newborn
individual's birth date, in which case a TRUE LowFrequency would arguably be more
appropriate. So why have these special cases at
all?
The primary aim is to accommodate individuals who didn't remain under continuous observation for very long. Short-lived infants, for example, may live fewer than 15 days and couldn't pass the 15/29 test, but they should nevertheless be considered residents during their brief lives. Similarly, if these infants do not live long enough to be censused more than 3 days, their brief lives would incorrectly appear to have occurred in a period of low frequency.
It remains true that an infrequently visited group
could be visited on a newborn individual's birth date,
and that it would be more accurate for that individual's
LowFrequency to be TRUE.
However, it is expected that short-lived individuals in
regularly observed groups will be much more common than
observed births in infrequently observed groups, so
these provisions have been created to preserve data
integrity for the former, admittedly at the expense of
the latter.
Regardless of the individual's Entrytype, the 15/29 test is always performed on the Entrydate. The test might not be used to obtain residency, but it is still needed to determine the bout's putative last date, and unless the Entrytype is B it is also needed to calculate the Entrydate's LowFrequency.
Once resident in a group, an individual remains resident until the end of the last consecutive 29-day window that passes the 15/29 test. The image below illustrates that there will inevitably be a number of days at the end of a residency period — at least 14 — that will not pass the 15/29 test.
Because of this, an individual's residency on any given date is not determined simply by whether or not they passed the 15/29 test. Once residency has been obtained, an alternative approach is needed to identify when a period of residency ends.
When the system iterates over a date on which the individual is already resident — when day 1 of the 29-day window is ≤ the putative last date of the current residency bout — the system will try to retain the individual’s residency. While the process of obtaining residency is about asking if the individual is resident on day 1, the process of retaining residency begins already knowing that to be true. Instead, the system attempts to extend the putative last date by asking if the individual is still resident on day 29.
Note
Even though retaining residency focuses on day 29 of a window, MEMBERS is only updated for day 1. The remaining rows in the window are updated in subsequent iterations when they become the “new” day 1.
Days 2-28 are neither ignored nor skipped; they are included in any 15/29 tests, they have already had a turn at being day 29 in earlier windows, and they will have a turn being day 1 in later windows.
While the individual continues to be present in the group with no absences from the resident group and no appearances in other groups, retaining residency is simple. In each consecutive window, the putative last date is updated to day 29 and the MEMBERS row for day 1 is updated to show that the individual was resident: Residency is R, LowFrequency is populated based on the density of data in that window, and GrpOfResidency is the row’s Supergroup. The system then iterates onward to the next 29-day window.
When the individual is not in the resident group on day 29, the system checks if the window passes the 15/29 test. If yes, the individual is away from the group but has not (yet) been away long enough to lose their residency. Lacking evidence to extend the putative last date, it is not changed. Regardless, the individual is still resident on day 1: the MEMBERS row with that Date is updated to indicate that the individual is resident: Residency is R, LowFrequency is populated based on the density of data in the window, and GrpOfResidency is populated as expected. The system then iterates onward to the next 29-day window.
When the individual is not in the resident group on
day 29 and the window fails the
15/29 test, the individual has
apparently been away long enough to
lose their residency. First, the system considers the
density of available data in the most-recent window that
did pass the test: if LowFrequency for that window is TRUE,
there may be an alternative explanation for the individual’s
absence, as discussed below in Retaining Residency with Low Frequency. If that
alternative is ruled out, or if the most-recent "passing"
window’s LowFrequency is FALSE,
the individual has certainly been away
long enough to lose their residency. The putative last date
becomes confirmed — no longer putative — and can
no longer be updated. When this first occurs, the confirmed
last date will necessarily be a date within the window and
later than day 1. Therefore, the individual is still
resident on day 1: the MEMBERS row with
that Date is updated to indicate
that the individual is resident: Residency is R, LowFrequency is populated with the value
used in the most-recent window that did pass the test, and
GrpOfResidency is populated as
expected. The system then iterates onward to the next
29-day window.
Once the residency’s last date is confirmed (not putative), the system won’t attempt to extend that date anymore and the process of retaining residency becomes a simple game of waiting for day 1 to become the last date. While day 1 is before or equal to the confirmed last date, the individual is certainly resident on day 1 and the related MEMBERS row is updated accordingly: Residency is R, LowFrequency is that of the most-recent window that did pass the 15/29 test, and GrpOfResidency is populated as expected. The system then iterates onward to the next 29-day window, unless day 1 is the confirmed last date. In that case, the residency bout is over and a corresponding row is added to the RESIDENCIES table. In the next 29-day window, there will be no residency to retain so the system will revert to attempting to obtain residency.
Before adding a new row to the RESIDENCIES table, the system categorizes how the residency ended. This information is recorded in the row's Finish_Status. See RESIDENCIES.Finish_Status for more information about possible values.
Note
While retaining residency, the system asks only whether the individual was in their resident group. When not in the resident group, the system does not pay attention to which group was visited. If an individual visits a group while resident elsewhere, the days of these visit(s) cannot count towards obtaining residency in the visited group later. The individual must lose residency before its visits to another group can count towards obtaining residency there.
When an individual is not regularly or frequently
censused, there may be periods of time when the individual
is not seen at all and is interpolated into the unknown
group (9.0). They will appear (in MEMBERS) to have left the group when (in
reality) they have not. Because of this, when LowFrequency is TRUE an individual’s
residency bouts might be extended across periods in which
their whereabouts are unknown.
When a resident of an infrequently censused group stops being seen and is interpolated into the unknown group (grp 9.0) long enough to fail a 15/29 test, their residency in that group may not actually end. The residency will extend through the “unknown” period in either of two situations. First, the individual retains residency if they are still in that group the next Date they are seen, provided that the individual was never censused “absent” from that group in the intervening time period. Second, the individual retains residency if the next Date that they are seen is the individual's Statdate and their Status is a Residency_Special_Case, as discussed below.
For every MEMBERS row in this
"unknown" period, the Residency
will be R, LowFrequency will be TRUE, and GrpOfResidency will be that of the
last window that passed the 15/29 test. If
that group ceases to
exist during this unknown period, the GrpOfResidency is whichever of that
group's daughter groups is first visited by the individual
after this "unknown" period ends.
Outside of the two aforementioned situations, the residency does not receive any special treatment and the residency bout ends as normal: on the last day the individual was seen (interpolated present) in the nonstudy group.
Caution
If a low-frequency group undergoes a fission or fusion during one of these "unknown" periods, the individual's presence in a parent group before the "unknown" period and in a daughter group afterward can only be recognized as the same group if the fission/fusion was in progress at the beginning or end (or both) of the "unknown" period. That is, 1) the last day before the "unknown" period or the first day after it must be during the part of a fission/fusion when the parent group(s) has not yet ceased to exist, or 2) the first day after the "unknown" period must be during the 28 days after the parent group(s) has ceased to exist, when the Delayed_Supergroup is the parent group. Outside of these circumstances, the individual is interpreted as being in two separate and entirely unrelated groups, and residency is not extended across the "unknown" period.
Sometimes, special treatment is needed when retaining residency and the last day of the window is the individual's Statdate. As the system approaches the end of available data for an individual, retaining residency may become disproportionately difficult. If the individual happens to be away from their grp of residency on their last day in MEMBERS, that single non-"present" day will cause them to prematurely lose their residency. In some cases that may be appropriate, but in other cases it does not reflect the demographic reality.
For example, if an individual is sick or injured near the end of their life and unable to keep up with the rest of their group, they eventually might fall so far behind that they are censused "alone" in their final few days. This would cause them to lose residency after their last present day in the group, apparently having dispersed from the group shortly before dying. But no, this hypothetical individual didn't socially leave their group; they just happened to be physically away from it at the time of their death. Their residency should end on their Statdate, not the last date present in the group.
In contrast, the end of an individual's data might occur because they dispersed from a study group to some unknown (never observed) other group. They were present in their group one day, seen alone the next, then never seen again. In that case, their residency will have ended because of a dispersal, so it would be quite appropriate for the individual to lose their residency as normal: on the last day they were present in the resident group.
In the above examples, the two different residency assignments made at the end of the individuals' MEMBERS data could be described with the same CENSUS data. The only difference between them: the manner in which they left the study population, i.e. their BIOGRAPH.Status. Thus, when attempting to retain residency in the days leading up to and including the individual's Statdate, the system allows some variation in the rules, depending on the individual's Status.
When the individual's Status is indicated in the STATUSES table as a Residency_Special_Case (when the
Residency_Special_Case is
TRUE), the MEMBERS row representing
the individual's Statdate is
treated as a sort of "wild card" in any
15/29 tests performed to retain residency.
That is, when attempting to retain residency and counting
the number of days that the individual was present in a
given group in a window, the Statdate will always count as
"present" in that given group. Thus, an individual who was
only briefly away will remain resident through their Statdate, but an individual who was
away long enough to fail a 15/29 test can
still lose their residency shortly before their
Statdate.
When an
individual is resident in a group with LowFrequency, this special case works
just a little differently and in one specific
circumstance. Suppose that an individual was last seen
present and resident in such a group, then was not seen
for an extended period of time until at last they were
found dead[207]. It is unclear if the individual was still a
resident of the group when they died, or if they had
dispersed to another group during that period of
nonobservation. The only datum available to address this
is the length of time between the individual's last Date in the resident group and the
individual's Statdate. When
the Statdate is more than
90 days after the
last Date in the group of
residency, it has been too long to make an inference about
the individual's residency and the special case does not
apply. When the Statdate is
90 or fewer days
after that last Date in the
group, the system infers that the individual had not left
their resident group, and the individual is eligible to
extend their residency through their Statdate. As in other cases where
low frequency residency is extended through "unknown"
periods, the individual's whereabouts must truly be
unknown during those final
90 or fewer days;
all MEMBERS rows in that time —
including that of the Statdate
— must have a Grp of
9.0, and the individual must
have no "absent" CENSUS rows in the
group of residency.
The "wild card" status for the Statdate risks allowing a narrow opportunity for an individual to pass a 15/29 test that the individual should have lost. If the individual is not in their resident group on their Statdate, and the 28 preceding days are evenly divided between the resident group and anywhere else (14 days in the group, 14 days in any other groups), then the individual has been away too long, and loses their residency. The "wild card" status in a residency special case does not extend the residency through the Statdate.
Individuals with any other Status are not forbidden from being resident through their Statdate. They simply must remain physically present in the group to retain their residency, as usual.
These tables all record females' sexual cycle states on a day-by-day basis, and provide daily measures of the number of days each female has been in and will remain in the given state. REPSTATS provides the broad overview and the remainder of the tables supply detail on the days REPSTATS indicates that the females are cycling. The day-by-day nature of these tables makes it easy to correlate reproductive cycle information with other events.
CYCGAPDAYS is something of an exception, in that it records days during which females are not under observation (according to a very specific definition.) It is included in this section because it exists to aid reproductive state tracking.
A day-by-day record indicating which days a female is not under observation. The definition of “not under observation” is that of CYCGAPS, see that table for more information. Contains one row per female per day during which the female is not under regular, continuous, observation.
Caution
Because the CYCGAPDAYS table primary purpose is to support the Babase system in it's validation and automatic analysis of the sexual cycle data an individual's last CYCGAPDAYS date is after the the BIOGRAPH.Statdate, should observation of the individual cease and not resume. This allows for easy determination of where there are gaps in observation and where automatic Mdates, which may occur after the individual's Statdate, must be generated.
This table is automatically constructed from the CYCGAPS table. It may not be manually maintained.
The female that is not observed. The three-letter code that identifies the individual's row in the BIOGRAPH table. There will always be a row in BIOGRAPH for the individual identified here.
This column may not be NULL.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
A day-by-day record of the details of the females'
cycles -- whether menses-follicular, swelling-follicular,
ovulating, or luteal. Contains one row per female per day,
for those days in REPSTATS for
which the REPSTATS State is C
(cycling.)
A female has rows in CYCSTATS whenever cycling; there are no CYCSTATS rows when a female is pregnant or lactating. Likewise there are no CYCSTATS rows when there are gaps in the observational record. (See CYCGAPS.) See the REPSTATS table for further detail as to exactly when a female is considered to be cycling, and for important cautions. See the description of the Din and Dr columns below for further information on how sexual cycles are recorded when there missing sexual cycle transition markers due to cessation of observation.
Caution
REPSTATS may show a female to be
cycling even when there are no rows in CYCSTATS for the
dates in question. This occurs when there are no CYCPOINTS during a period of observation. This
can only occur for females without a MATUREDATES.Mstatus of
O when observation ceases before
the first observed sexual cycle transition event.
The system will issue a warning when REPSTATS indicates a female is cycling but there is no row in CYCSTATS for the day in question.
Caution
Females may become turgesent (have a Tdate) on the day
they are in menses (Mdate). As CYCSTATS has a 1 day
resolution and, essentially, these females are in menses for
less than a day, when this happens CYCSTATS will not show
any days in menses (State is M)
for these cycles even though the cycle has an Mdate row in
CYCPOINTS.
Similarly, when there are less than 6 days between an
Mdate and the following Ddate a cycle will have no days in
the swelling-follicular state (State is
S).
Caution
When the last date of a
S (Swelling-follicular) cycle
state is not known[208], that is, a cycle has no Ddate due to
cessation of observation, death, delay in data entry, or
whatever other reason, two problems arise that will, unless
accounted for, adversely affect sexual cycling
analysis. First, the O
(Ovulating) state will not occur because the transition
between S and the
O state is determined by the
following Ddate[209], which does not exist. Second, because the
O state cannot be
calculated, the S state may
be erroneously lengthy; days when the female is actually in
the ovulating state may be marked with a
S rather than an
O and these rows will have
an incorrect Din (days into state) values.
Rather than omit the accurate
S rows along with the
inaccurate, the Babase designers chose to include all
available data to accommodate those analysis that do not
distinguish between the S
(Swelling-follicular) state and the
O (Ovulating) state. The
Babase user is expected to know the conditions under which
various data may be used.
Note
In the case of an individual that has ceased cycling due to pathology or old age, and whose last cycle did not end in pregnancy, the final CYCSTATS rows will have a State of D and an unusually long duration, with the individual's date of death being the last day of the cycle.
The sum of Dins and Dr is always the total number of days the cycle spent in the state.
Warning
Babase does not populate this table automatically, although we would like it to do so. The rebuild_all_cycstats() or rebuild_cycstats() programs must be manually executed to ensure the content of this table corresponds with that of the rest of the database.
Users cannot directly manipulate the table's data.
Categorizes the period within the reproductive cycle. Legal values are:
| Code | Mnemonic | Description |
|---|---|---|
M | Menses-follicular | the Mdate (onset of menses) to the day before the Tdate (turgesence onset) (inclusive of endpoints) |
S | Swelling-follicular | the Tdate through 6 days before the Ddate (deturgesence onset) (inclusive of endpoints) |
O | Ovulating | from 5 days before the Ddate through one day before the Ddate (inclusive of endpoints) |
D | Deturgesence | luteal -- from the Ddate through the day before the Mdate (inclusive of endpoints) |
The number of days since the state started. The first
day of the state has a value of 1, the
next a value of 2, etc.
This column is NULL when the system cannot determine
when the state began. This happens when the cycle's starting
date occurs during a period when the individual is not under
regular observation. (See CYCGAPS.)
The number of days remaining in the state. The last
day of the state has a value of 0, the
next to last day a value of 1,
etc.
This column is NULL when the system cannot determine
when the state ends. This occurs when the end of the cycle
was not observed, either because the individual is alive and
additional observations have not yet been entered into
Babase or due to cessation of regular observation. (See
CYCGAPS.) It also occurs
when the individual dies while cycling as it is not known
when the state would have ended.
The Cpid of the CYCPOINTS row recording the sexual cycle
transition event that started the state. NULL when
there is no such row. See REPSTATS.Dins for
further detail.
The Cpids value of CYCSTATS rows with a State of
O (Ovulating) reference a
Tdate (Code of
T) CYCPOINTS row, even though the Tdate is not
(usually) the first ovulation date. This is because the
Tdate, if it exists, if the Cpids is not NULL, is the
sexual cycle transition event which precedes the
ovulation. The Dins column should be subtracted from the
Date column to find the first day of ovulation.
The Cpid of the CYCPOINTS row recording the sexual cycle
transition event that ended the state. NULL when there
is no such row. See REPSTATS.Dr for further detail.
The Cpide value of CYCSTATS rows with a State of
S (Swelling-follicular)
reference a Ddate (Code of
D) CYCPOINTS row, even though the Ddate is not
the day after the last day of the swelling-follicular
state. This is because the Ddate, if it exists, if the
Cpide is not NULL, is the sexual cycle transition event
which follows the swelling-follicular state. The Dr
column should be added to the Date column to find the last
day of the swelling-follicular state.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
A day-by-day record of the number of days since the
previous Mdate/until the next Ddate. Contains one row per
female per day, for those days in REPSTATS for which the REPSTATS State is C
(cycling), for those days between the cycle's Mdate and Ddate,
inclusive of the Mdate but exclusive of the Ddate. This table
contains rows whenever there are rows on CYCSTATS. See the CYCSTATS
documentation for further details and the REPSTATS documentation for details and
cautions.
When there is no prior Mdate, due to pregnancy,
menarche, or resumption of observation, the Dini column is NULL. However, the
corresponding row in the REPSTATS table contains what may be
a relevant Din value.
Note
In the case of an individual that has ceased cycling due to pathology or old age, that individual's final Mdate to Ddate interval will have a long duration, with the individual's date of death being the last day of the interval.
The sum of Dini and Dr is always the total number of days counting[210]from the cycle's Mdate up to[211] its Ddate.
Warning
Babase does not populate this table automatically, although we would like it to do so. The rebuild_all_mdintervals() or rebuild_mdintervals() programs must be manually executed to ensure the content of this table corresponds with that of the rest of the database.
Users cannot directly manipulate the table's data.
The row records the number of days until the cycle's Ddate/from the cycle's Mdate relative to this day.
The number of days into the interval. The first day
of the interval, the Mdate at the beginning of the
interval, has a value of 1, the next
day a value of 2, etc.
This column is NULL when there is no Mdate at the
beginning of the interval. This occurs when the cycle is
the female's first cycle, as there is no menses to begin the
cycle, and likewise for the first cycle after pregnancy. The
cycle's Mdate is also unknown when it occurs during a period
when the individual is not under regular observation. (See
CYCGAPS.)
The number of days remaining in the interval -- days
to, but not including, the Ddate that ends the interval.
The last day of the interval, the day before the Ddate
that ends the interval, has a value of
0, the day before that a value of
1, etc.
This column is NULL when there is no next Ddate,
either because the individual is alive and additional
observations have not yet been entered into Babase or due to
cessation of regular observation. (See CYCGAPS.) It can also occur when
an individual dies.
The Cpid of the CYCPOINTS row recording the starting Mdate.
NULL when there is no such row, when the interval occurs
at the beginning of a period of continuous observation
(see CYCGAPS), after a pregnancy, or at
menarche.
The Cpid of the CYCPOINTS row recording the ending Ddate.
NULL when there is no such row, when the interval occurs
at the end of a period of continuous observation (see
CYCGAPS) or the point of cessation of
data entry.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
A day-by-day record of the number of days since the
previous/until the next Mdate. Contains one row per female
per day, for those days in REPSTATS for which the REPSTATS State is C
(cycling). The Mdate-to-Mdate interval includes the Mdate at
the beginning of the interval but does not include the Mdate
at the end of the interval[212]. This table contains rows whenever there are
rows in CYCSTATS. See the CYCSTATS documentation for further details and the
REPSTATS documentation for details and
cautions.
When there is no previous Mdate, due to pregnancy,
menarche, or resumption of observation, the Dini column is NULL. However, the
corresponding row in the REPSTATS table contains what may be
a relevant Din value.
When there is no subsequent Mdate due to pregnancy,
death, interruption of observation, or cessation of data
entry, the Dr value is NULL.
When there is no subsequent Mdate due to pregnancy what may be
a relevant Dr value can be found in the REPSTATS table.
Note
In the case of an individual that has ceased cycling due to pathology or old age, that individual's final Mdate to Mdate interval will have a long duration, with the individual's date of death being the last day of the interval.
The sum of Dini and Dr is always the total number of days between Mdates.
Warning
Babase does not populate this table automatically, although we would like it to do so. The rebuild_all_mmintervals() or rebuild_mmintervals() programs must be manually executed to ensure the content of this table corresponds with that of the rest of the database.
Users cannot directly manipulate the table's data.
The number of days into the interval. The first day
of the interval, the Mdate at the beginning of the
interval, has a value of 1, the next
day a value of 2, etc.
This column is NULL when there is no Mdate at the
beginning of the interval. This occurs when the cycle is
the female's first cycle, as there is no menses to begin the
cycle, and likewise for the first cycle after pregnancy. The
cycle's Mdate is also unknown when it occurs during a period
when the individual is not under regular observation. (See
CYCGAPS.)
The number of days remaining in the interval -- days
until the Mdate which follows the interval[213]. The last day of the interval, the day
before a Mdate that comprises the end of the interval, has
a value of 0, the day before that a
value of 1, etc.
This column is NULL when there is no next Mdate,
either because the individual is alive and additional
observations have not yet been entered into Babase or due to
cessation of regular observation. (See CYCGAPS.) It can also occur when
an individual dies while cycling as it is not known when the
state would have ended.
The Cpid of the CYCPOINTS row recording the earlier Mdate.
NULL when there is no such row, when the interval occurs
at the beginning of a period of continuous observation
(see CYCGAPS), after a pregnancy, or at
menarche.
The Cpid of the CYCPOINTS row recording the later Mdate.
NULL when there is no such row, when the interval occurs
at the end of a period of continuous observation (see
CYCGAPS) or ends in pregnancy.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
A day-by-day record indicating whether a female is pregnant, lactating, or cycling. Contains one row per female per day for every day during intervals of continuous observation from date of menarche through date of death (inclusive). When menarche is unobserved then REPSTATS rows begin on a beginning of observation date.[214] Likewise, the cessation or resumption of observation interrupts or resumes the contiguous series of the female's REPSTATS' dates. (See CYCGAPS.) While the individual is alive[215], and under observation, the last date is either the BIOGRAPH.Statdate or the last recorded sexual cycle endpoint, which ever is later. When the individual is not alive, but was under observation until death, the last date is the female's Statdate.
Warning
Because Babase generates REPSTATS rows ending, at minimum, with females' Statdates, the data entry staff should enter sexual cycle information (CYCPOINTS and CYCGAPS) for a time interval before entering demographic information (CENSUS, BIOGRAPH Statdate and Status) for that interval, otherwise Babase may continue a particular reproductive state to the Statdate when there are reproductive data to the contrary yet to be entered.
Caution
Babase assumes individuals are under continuous observation. If there is no record of a gap in observation (see CYCGAPS), the entire interval between the onset of menarche (Matured) and the first recorded sexual cycling event (CYCPOINTS) is included in the individual's first reproductive state interval in REPSTATS and possibly in CYCSTATS, MMINTERVALS, and MDINTERVALS as well.
Note
Because of gaps in the observational record, some sexual
cycles may not be recorded, or may be partially recorded. In
these cases the Dins and Dr columns are NULL. (See
below.)
The sum of Dins and Dr is always the total number of days spent in the state.[216]
Warning
Babase does not populate this table automatically, although we would like it to do so. The rebuild_all_repstats() or rebuild_repstats() programs must be manually executed to ensure the content of this table corresponds with that of the rest of the database.
Users cannot directly manipulate the table's data.
See CYCSTATS, MMINTERVALS, and MDINTERVALS for more fertility detail.
General reproductive state of the female on the given Date. The legal values are:
| Code | Mnemonic | Description |
|---|---|---|
C | Cycling | From (including) the Tdate (turgesence onset) up to (but not including) the Ddate of the onset of pregnancy. |
P | Pregnant | From (including) the Ddate (deturgesence onset) up to (but not including) the end-of-pregnancy date, i.e., the date that the female experiences an infant birth, experiences a spontaneous abortion, or dies. |
L | Lactating | Postpartum amenorrhea. From (including) the end-of-pregnancy date to (but not including) the next Tdate. |
Caution
The above definition of pregnant means that on the conception date the mother is in a pregnant state, even though the conception date is a Ddate and the Ddate has a cycle (a Cid on CYCPOINTS).
Note
REPSTATS does not keep track of whether a female's
cycles are “normal”; it simply forces
females into one of these three states. Individuals who
have ceased cycling or have irregular cycles due to
pathology or old age have a state of
C, or possibly
L if the last cycle
resulted in a pregnancy.
Any of the above states may start late or end early in the event of gaps in observation. (See CYCGAPS.)
The number of days since the state started. The first
day of the state has a value of 1, the
next a value of 2, etc.
This column is NULL when the system cannot determine
when the state began. This occurs when the beginning of the
reproductive state occurs during a period when the
individual is not under regular observation (see CYCGAPS) or when an individual's
sexual maturity date is not also a Tdate (see MATUREDATES).
The number of days remaining in the state. The last
day of the state has a value of 0, the
next to last day a value of 1,
etc.
This column is NULL when the system cannot determine
when the state ends. This occurs when the end of the
reproductive state was not observed, either because the
individual is alive and additional observations have not yet
been entered into Babase, or due to cessation of regular
observation. (See CYCGAPS.)
It also occurs when the individual dies, as it is not known
when the state would have ended.
The Pid of the
pregnancy associated with the state. This value must be
present when the State is P
(Pregnant) or L
(Lactating). There is also a Pid value for those
C (Cycling) states that end
in pregnancy; this will apply to the majority of the
C states, as the only other
way to exit the C state is
death or cessation of observation.
The timestamp range during which this row's data are considered valid. See The Sys_Period Column for more information.
Sexual cycles (CYCLES) are defined by Mdate, Ddate, and Tdate sexual cycle transition events. CYCLES should be created and destroyed in correspondence with Mdate, Tdates, and Ddates. But Babase contains other information related to sexual cycles, most obviously sexskin swelling This section describes how this information is related to specific sexual cycles.[217]
Note
The determination of when a new sexual cycle starts is, because by definition a cycle is a periodicity with no start and no end, arbitrary[218], as then is the determination of which cycle to associate various data with. The method used by Babase was chosen for its simplicity and its ability to be consistently applied to all sorts of cycle related data. It may lead to what may be non-intuitive results. As with all things Babase, users must take care to familiarize themselves with the intricacies[219] of the system, and the data.
Babase uses the date of the measurement, of whatever data, sexskin swelling, PCS color, etc., to determine which sexual cycle the measurement should be associated with. Dates are assigned to cycles by virtue of falling in the interval each cycle spans, each cycle starting with an Mdate and continuing through the day before the next Mdate; although cycles can be “cut off” by cessation, or initiation, of observation. The following method implements these policies and can be used as a guide when there are questions as to the specifics:[220]
Relate the measurement to the cycle of the Mdate, Tdate, or Ddate that falls on the date of the measurement or is the latest Mdate, Tdate, or Ddate preceding the measurement, so long as there is no gap in observation between the measurement date and Mdate, Tdate, or Ddate. If there is no such Mdate, Tdate, or Ddate due to gaps in observation or simple lack of data then relate the measurement to the cycle of the earliest Tdate or Ddate that follows the measurement but is not separated from the measurement by a gap in observation or an intervening Mdate. If there is no such Tdate or Ddate then the measurement may not be recorded in Babase.
Warning
Because there are conditions under which sexual cycle related data may not be recorded in Babase, and, as a rule, Babase does not automatically delete data, Babase will not permit some orderings of data maintenance operations. For example, Babase will not allow a gap in observation to be inserted after a female's last Ddate but before her last sexual swelling date because this would require removal of the sexual swelling information. An alternate ordering of the operations resulting in identical database content is required. In the above example either the sexual swelling data must be deleted or subsequent Mdates, Tdates, or Ddates must be entered before the gap in observation may be entered.
Note
This section describes how Babase automatically re-computes the sequence numbers used within various tables to give a timewise ordering to rows that would not otherwise have an ordering. The columns that hold the sequence values have names that vary by table. The following description uses the generic column name of “Seq” when referring to the name of the column that holds the sequential numbering.
The system automatically re-computes Seq values to
ensure that they are contiguous and begin with
1. Seq may be NULL when the row is
first inserted, in which case the system will automatically
assign the next available sequence number. Changing a
sequence number to match one that already exists (for, e.g.,
a given darting), or inserting a new row having a sequence
number equal to that of an existing row (for, e.g., a given
darting) causes the sequence number of the unchanged row to
be incremented and the recomputation of subsequent sequence
values. E.g. starting with rows A, B, C, and D having Seq
values of 1, 2,
3, and 4 respectively,
changing the Seq value of row D to 2
automatically changes the Seq values of rows B and C,
increasing them by one. The result is that the new ordering
of the rows by sequence number becomes: A, D, C, B.
Deleting a row recomputes the sequence numbers of the
remaining rows in a corresponding fashion.
Caution
Updating a row to increment the sequence value by
1 will do nothing[221]. Performing such an operation creates a
“gap” in the sequence which is then
“filled” by decrementing the sequence numbers
of all the rows “above the gap”, including
the row that the original update incremented.
Likewise, updating the Seq column in a way that assigns Seq numbers past the “end” of the sequence results not in the user-specified Seq values but rather in Seq values that are re-computed so as to maintain contiguity.
Warning
A single UPDATE statement that relies
on automatic resequencing to eliminate more than one
duplicate Seq (per, e.g., a given darting) produces
indeterminate results.[222] For example given rows A, B, C, and D, with
Seq values of 1, 2,
3, and 4 respectively.
One UPDATE statement that changes the Seq of A
to 3 and B to 4 will
result in an indeterminate ordering.[223]
The system will report an error when the Seq values of
inserted rows would create non-contiguous Seq values or a
sequence that does not begin with
1.[224]
CYCPOINTS is special in that the
presence of a Ddate row can trigger the automatic generation of
a Mdate row 13 days later. Automatically generated Mdates are
distinguished by having a CYCPOINTS.Source of A.
As Ddate rows are inserted, updated, or deleted Babase makes
appropriate changes to ensure that automatically generated Mdate
rows exist on the 13th day following a qualified Ddate. The
exception is when a Tdate follows a Ddate by less than 13 days
(and there are no intervening gaps in observation.) In this
case the automatically generated Mdate will have the Tdate's
date and be less than 13 days after the previous Ddate.
An Mdate will be generated from a Ddate when all of the following conditions are met:
Either there is no Mdate in the cycle following the Ddate's cycle or there is a gap in observation between the Ddate's cycle and the following cycle.
The Ddate is not the start of a pregnancy, its Cpid does not appear as a Conceive value on the PREGS table.
Observation proceeds without a gap for at least 13 days following the Ddate, or up to the Tdate immediately following the Ddate, which ever comes first,
The Ddate is not estimated. (Source is not
E)The individual is alive (BIOGRAPH.Status is
0) on the automatic Mdate.[225]
A Mdate automatically generated from a Ddate will be
removed when any of the above conditions are no longer met, or
when another Mdate is automatically generated for the
Ddate.[226]More precisely, it is not a Mdate automatically
generated from a Ddate that will be removed but rather any Mdate
will be removed that has a Source of
A, and that post-dates the Ddate, and
that has no Mdates, Tdates, or Ddates, or periods of no
observation (see CYCGAPS) on the interval
between the Ddate and the “automatic” Mdate.
Babase cannot distinguish manually entered Mdates with a Source of A
from automatically generated Mdates. Therefore it is not just
automatically generated Mdates that will be removed.
Automatically assigned Mdates, those with a
Cycpoints-Source of A, have NULL
Edates and Ldates.
[168] Ideally, the interpolation algorithim would be written to ensure that individuals cannot be interpolated into groups that did not exist on the indicated Date. If this were so, a separate check in MEMBERS wouldn't be needed. However, this modification to the code is more complicated than one might expect. For various practical reasons, it's ideal to enforce this "group must exist on this date" rule "on commit" of an SQL transaction. In constrast, the interpolation algorithim operates independently of transactions (it was written before the technology to enforce anything "on commit" existed in PostgreSQL). Effectively incorporating this validation of the Date into interpolation will require rewriting interpolation to work in transactions. This will likely be a substantial rewrite, so for now, interpolation and Date validation are performed separately.
[169] To be perfectly clear, the residency status (MEMBERS.Residency) is that of the social group (MEMBERS.GrpOfResidency).
[171] But an absence and a presence recorded on the same day count only as a single day of censusing.
[172] For example, when the row for an individual at rank 1 is inserted, the Ags_Density, Ags_Reversals, and Ags_Expected can't yet be calculated accurately because the rows for individuals ranked 2 and onward have not yet been added.
[173] Note that the requirement that ranks be contiguous means that in order to change an existing ranking the ranks must first be deleted, from highest numbered rank to lowest, and then the new ranking re-created, from lowest numbered rank to highest.
[174] ...which only happens with adult male ranks.
[175] Because in all current (as of this writing) laboratory protocols, methanol and solid-phase extractions are always in the same series, and ethanol extraction is always part of another.
[176] If this restriction is ever lifted, the hormone-specific views (e.g. ESTROGENS) will not be guaranteed to be one-row-per-sample. This isn't necessarily a problem, but it's a downstream effect that may not be immediately obvious and seems worth noting.
[177] For other examples of this, see the NUCACID_LOCAL_IDS and TISSUE_LOCAL_IDS tables, and the WP_REPORTS.WId column).
[178] Usually the olive baboon, Papio anubis.
[179] For discussion in this table, we use the term, "confidence interval" generally. It may not necessarily be an actual "confidence interval" as a statistician would use that term. The confidences recorded in this table may actually be another kind of interval, or another kind of confidence.
[180] At this time only DEMOG, the demography notes table, contributes to CENSUS any information regarding group membership.
[181] Sometimes, when demography information is added into other tables, CENSUS rows are altered rather than removed. Likewise, CENSUS rows are removed (or altered as necessary) when demography information is removed from other tables.
[183] This is the one exception, if you wish to consider it so, to the rule that an individual cannot be censused both present and absent in the same group on the same day.
[184] The “same group” condition is one that must be met whenever interpolation examines intervals between presence and absence.
[185] As the individual is alive, every census that post-dates the individual's Statdate must record an absence, else the Statdate would be adjusted to reflect the date of last census.
[186] This is a heuristic. While it should work well enough most of the time the Babase user must be aware of the pitfalls in this approach. These are explained below.
[187] Without this restriction interpolation would have to insert rows forever, placing the individual in the unknown group off into the indefinite future.
[188] Notice that interpolation does not bother analyzing absences, such as the last-most, that are not neighbor to censuses.
[189] Note that the intervals spoken of here are always anchored at one end by a census that finds an individual in a group. Each such census can therefore have 2 intervals associated with it, one of the days preceding the census date and one containing the following days. These intervals can then appear in the diagrams as single lines that contain a census date. It is important to remember that there are really 2 intervals depicted; one line that ends on the date of the census and another that begins at that point.
[190] As locating censuses are interpolated individually the figure could diagram the intervals associated with each census separately, as in Figure 4.14, work out group membership from that, and then combine the results; the outcome would be unaffected. The chosen presentation form allows the interval endpoints to “match up” in a revealing fashion. As an exercise the reader should prove to himself that the intervals associated with each locating census are accurately depicted, and that the order in which locating censuses are interpolated does indeed make no difference.
[191] Figure 4.14: “A Closer Look at Intervals” makes clear that it is not necessary to show these intervals. By definition, the omitted intervals will always be longer than the “halfway to census” interval of the census being interpolated. As the shorter interval is the one used the longer may be ignored.
[192] When there are two intervals. When there's no “absence” interval the “Used:” line shows the “presence” interval.
[193] The proper term is “The Glorious Interpolation Procedure”, but we don't tell this to just anybody.
[195] It might be better if interpolation did not
interpolate at all on those intervals between
interpolating censuses that contain a non-interpolating
census[195] -- if it put the individual in the unknown group, with an Interp of 0 and
an Origin of NULL whenever
there was no locating census. However, this
could easily cause problems because interpolation has
always worked as the body of this document describes.
Although these situations are not supposed to occur,
it is likely the data contains such situations and
changes should not be made to interpolation which
break the database.
[195195] I have not thought this through. At first glance it seems the code would be simpler, but perhaps not. And the effect on data analysis is unclear. It is probably best to adopt one of the solutions presented in the note below.
[197] Although in this example we “count
up” traversing the timeline from left to right,
had the N census
had been closer to the right side of the diagram than
the left we would be “counting up” the
interval by traversing the timeline in the opposite
direction, from right to left.
[198] The same method is used to compute Interp values when interpolation uses The 3 Interpolation Intervals, above.
[199] This “same group” criteria corresponds with the criteria found in The Halfway to Absence Interval.
[200] Interp is fixed at
0 over the portion of The Halfway to Census Interval that was truncated in the
preceding paragraph. Effectively, as MEMBERS Interp
counts up with increasing distance from the
interpolating census, the count is fixed at NULL
upon encountering a non-interpolating census until the
point is reached at which counting back down to the
next interpolating census begins, at which point the
count downward resumes as though never
interrupted.
[201] This is examined in detail in Interpolation at the Statdate.
[202] From this discussion, it's tempting to conclude that residency can never be obtained/retained in a group before it becomes Permanent, but that would be an overgeneralization. Individuals can become resident in a non-permanent group if there is no parent group to be resident in. That is, individuals can be residents of a group before its Permanent date if the group has no From_group and does not exist as another group's To_group in the GROUPS table.
[203] Note that this is only an issue after a fission. After a fusion, there are not multiple daughter groups to switch between.
[204] I.e. if group 1 divided into groups 2 and 3, the system would choose group 2 because it comes before 3 when ordered numerically.
[205] Which would only happen if the group ceased to exist and has no daughter group(s).
[206] In practice, individuals with those Entrytypes probably shouldn’t be in either of those groups on their Entrydate anyway, but there are no rules that explicitly forbid it.
[207] For individuals who have been fitted with a radio collar this is not unusual.
[208] A circumstance easily detected because Dr (days
remaining in state) is NULL.
[209] See the information on the calculation of the
S (Swelling, follicular)
and the O (Ovulating)
states below.
[210] Starting with 1.
[211] but not including
[212] which is part of the next Mdate-toMdate interval
[213] And the presence of which ends the interval.
[214] For females with a MATUREDATES.Mstatus that is not
O (On), this is the later of
MATUREDATES.Matured and the start of
observation according to CYCGAPS, as
expected.
[216] Or NULL, when either column is NULL, as adding a
NULL to anything results in NULL.
[217] For information on how Mdates, Tdates, and Ddates are aggregated into sexual cycles see both the CYCLES and the CYCPOINTS documentation.
[218] The decision to define a cycle as starting with an Mdate and ending in a Ddate is traditional, and yet not entirely sensible as the first cycle at menarche will assuredly not have a Mdate. In addition should an individual cease cycling due to pathology or old age the last cycle will not have a Tdate or Ddate. A definition of cycle that more closely parallels the life of an egg, starting with Tdate and ending with Mdates, would seem to make the most sense.
[219] Babase doesn't have quirks, it has intricacies. This will be on the midterm.
[221] Well, it will waste some electrons.
[222] Technically an UPDATE statement
that, in the absence of any triggers, would result in
more than one Seq value (for any given, e.g., Dartid)
within a contiguous series of Seq values as examined
after the UPDATE is an UPDATE that results in an
indeterminate ordering (within the, e.g., Dartid).
However in the future this behavior may change such
that any duplication of Seq values, not just those
within a contiguous series of Seq values, may result
in an indeterminate ordering.
[223] The problem is that duplicate Seq values are eliminated on a row by row basis. When more than one duplicate exists (per, e.g., a given darting) the order in which duplicates are eliminated matters. But when 2 or more duplicates are created at once there is no way to control the order in which the system processes the removal of duplicates.
[224] This is done so that data entry errors are not “invisibly corrected” under the assumption that when a Seq value is deliberately assigned to a new row that there is a reason for the assignment. Updates that make the Seq numbers “too large”, that would create gaps in the sequence if not corrected, do not result in errors but are automatically fixed. The latter behavior could be considered a bug; one to be fixed if it ever causes a problem.
[225] This means that automatic Mdates may occur after an individual's Statdate, so long as the individual is alive.
[226] So, there does not have to be a special rule to change the date of automatically generated Mdates in response to changes in the Ddates that generated them. Altering the Ddate creates a new Mdate, and in response the old Mdate is removed.
Table of Contents
- General Support Tables
- Group Membership and Life Events
- BSTATUSES (Birth Accuracy Indicators)
- CONFIDENCES (death cause (nature and agent), dispersal, and matgrp Confidence levels)
- DADS_COMPLETENESS (Completeness Scores in Paternity Assignments)
- DADS_EVIDENCE_TERMS (Types of data in paternity analyses)
- DADS_MISMATCHES (Types of Genetic Mismatches)
- DCAUSES (Causes of Death)
- DEMOG_REFERENCES (Demography Note References)
- DEATHNATURES (Natures of Death Causes)
- ENTRYTYPES (Categories of Entry to Study Population)
- MSTATUSES (Maturity Marker Statuses)
- RNKTYPES (Ranking Categories)
- STATUSES (Indicators of Record and Baboon Vividity)
- Physical Traits
- Social And Multiparty Interactions
- ACTIVITIES
- ACTS (Interaction Types)
- DATA_STRUCTURES (Data structures produced by Psion devices)
- CONTEXT_TYPES (multiparty Interaction Context Categories)
- FOODCODES (Food item Codes)
- FOODTYPES (Food Types)
- KIDCONTACTS (spatial relationship between mother and infant)
- MPIACTS (Multiparty Interaction Types)
- NCODES (Neighbor classifications)
- PARTUNKS (problem identifying a multiparty interaction participant)
- POSTURES
- PROGRAMIDS (Program used on the device)
- SAMPLES_COLLECTION_SYSTEMS
- SETUPIDS (Setup files used in a data collection program)
- STYPES (Focal Sample Types)
- STYPES_ACTIVITIES (Activity values that are used with each SType)
- STYPES_NCODES (Ncodes that are used with each SType)
- STYPES_POSTURES (Postures that are used with each SType)
- SUCKLES (infant suckling activity)
- Sexual Cycles and The Sexual Cycle Day-By-Day Tables
- Darting
- DART_SAMPLE_CATS (Darting Sample Categories)
- DART_SAMPLE_TYPES (Sample Types)
- DRUGS (darting anesthetics)
- LYMPHSTATES (Lymph node conditions)
- PARASITES (Parasites and their indicators)
- TCONDITIONS (Tooth Conditions)
- TICKSTATUSES (parasite count classifications)
- TOOTHCODES (kinds of teeth)
- TOOTHSITES (Locations of deciduous or adult teeth)
- TSTATES (State of Tooth existence)
- Inventory
- INSTITUTIONS
- MISID_STATUSES (MISIDentification STATUSES)
- NUCACID_CONC_METHODS (NUCleic ACID CONCentration quantification METHODS)
- NUCACID_CONC_UNITS (NUCleic ACID CONCentration quantification UNITS)
- NUCACID_CREATION_METHODS (NUCleic ACID CREATION METHODS)
- NUCACID_TYPES (NUCleic ACID TYPES)
- STORAGE_MEDIA
- TISSUE_TYPES
- SWERB Data
- ADCODES (SWERB Ascent and Descent relationships)
- PLACE_TYPES (codes for various landscape features)
- PREDATORS (codes for observed predators)
- SWERB_LOC_CONFIDENCES (SWERB Location Confidence Values)
- SWERB_LOC_STATUSES (SWERB Location Statuses)
- SWERB_TIME_SOURCES (SWERB Time Sources)
- SWERB_XYSOURCES (SWERB Time Sources)
- Weather Data
- Metadata
The support tables are those tables that define various codes used as data values in other tables. They define the controlled vocabulary used elsewhere in the system. The formulation of the available vocabulary is, for the most part, up to the users of Babase. This provides a great deal of flexibility in the information Babase records without requiring any programmatic or other alteration to the Babase system itself. New code values can be added to the system and used in the data by adding new rows to the support tables. The system validates the new code values in the data tables against the rows of the support tables allowing new types of data to be recorded without requiring changes to the Babase system.
Caution
Some of the vocabulary in the support tables has special meaning to Babase. All values that have a special meaning to Babase are noted in each table's documentation. Care must be taken when making changes in these cases or Babase will break. See the Special Values section for further information.
Most support tables contain only two columns (not counting
the Sys_Period column): a
key or id column that usually has the same name as the column in
the tables for which the support table defines vocabulary, and a
column called Descr. The key column contains the valid code
values, and the Descr column contains a short description of the
code. Both the key column and the Descr column must contain
values that are unique among all the values of all the rows in the
respective column. Neither the key column nor the Descr column
may be NULL. Neither the key column nor the Descr column may be
empty, contain no characters. Neither the key column nor the
Descr column may contain nothing but spaces.
As with nearly every other table in Babase, every support table has a Sys_Period column that shows the range of time during which the row's data is considered valid. See The Sys_Period Column for more information.
Some support tables contain one or more additional columns. These are described in the section devoted to the table at hand.
These support tables are used throughout Babase.
The different parts of the body examined for ticks when darting. These are not necessarily mutually exclusive. If, e.g., ticks are at times counted on the left foreleg and at times counted on the inner left foreleg and the outer left foreleg then this table would contain 3 rows, one for the entire leg and one each for the inner and outer portions.
Each combination of Bodyside, Innerouter, and Bodyregion must be unique.
BODYPARTS defines values for TICKS.Bodypart and WP_AFFECTEDPARTS.Bodypart.
Whether the bodypart is on the left side, right side,
center, or unspecified/not applicable. The corresponding
values are L,
R,
C and
N.
Whether the bodypart is on the inner (anterior) part
of the body, the outer (posterior) part of the body, or
whether this is unspecified/not applicable. The
corresponding values are I,
O, and
N.
The code for the part of the body of which the given part is a component -- a BODYPARTS.Bpid value. This column allows the establishment of a hierarchical relationship between the different parts of the body.[227]
This column may not be NULL. Body part rows that represent the highest
level of aggregation should reference their own Bpid value.
Contains one row for each person involved with the creation of data that was generated via laboratory techniques and procedures. This is a separate list[228] from that of the personnel involved with the observing of data, who are recorded in OBSERVERS.
Only the Babase administrator can create LAB_PERSONNEL rows with Initials values that the NUCACIDS view cannot reliably distinguish. See the portion of the NUCACIDS documentation which describes the ability of the view to distinguish one creator from another. Unless it can be assured that such indistinguishable creators will never simultaneously create nucleic acid samples then creating such Initials is not recommended.
LAB_PERSONNEL defines vocabulary for the HYBRIDGENE_ANALYSES.Analyzed_By, NUCACID_CREATORS.Creator, and WBC_COUNTS.Counted_By columns.
Initials
The initials of the person. This is used to uniquely identify the person, so may not be the person's actual initials if there is ever a conflict with a pre-existing value.
This column may not be empty, it must contain characters, and it must contain at least one non-whitespace character.
The person's real name.
This column may not be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
Textual remarks regarding when the person was doing lab work. Usually a date range.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
Any miscellaneous notes about the person. For example you may wish to record that the person is John Smith the graduate student, not John Smith the President of Kenya who asked to help with a DNA extraction and actually did so one day.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
Contains one row for each person who records data that was seen/witnessed/observed. This table may include people who assist the data collection process, whether in our out of the field and whether or not their initials appear in those database columns for which the OBSERVERS table provides a validation vocabulary because the initials of all these people may appear in paper or unvalidated electronic records.
OBSERVERS is unusual in that, in some sense, it has two key columns: Initials and OldGPSInititals. Which key is used in the field depends upon the data collection protocols. When entered into Babase all OldGPSInititals values are translated into their respective Initials values, so it is the Initials values that Babase always uses to reference the individual.
Only the Babase administrator can create OBSERVERS rows with Initials values that the SWERB_UPLOAD view cannot reliably distinguish. See the portion of the SWERB_UPLOAD documentation which describes the ability of the view to distinguish one observer from another. Unless it can be assured that such indistinguishable observers will never simultaneously collect SWERB data then creating such Initials is not recommended.
Likewise, only the Babase administrator can create OBSERVERS rows with Initials values that the WP_REPORTS_OBSERVERS view cannot reliably distinguish. See the portion of the WP_REPORTS_OBSERVERS documentation which describes the ability of the view to distinguish one observer from another. Unless it can be assured that such indistinguishable observers will never simultaneously collect Wounds and Pathologies data then creating such Initials is not recommended.
OBSERVERS defines vocabulary for INTERACT_DATA.Observer, SAMPLES.Observer, WREADINGS.WRperson, RGSETUPS.RGSPerson, CROWNRUMPS.CRobserver, CHESTS.Chobserver, ULNAS.Ulobserver, HUMERUSES.Huobserver, and SWERB_OBSERVERS.Observer.
Initials
The initials of the person. This is used to uniquely identify the person, so may not be the person's actual initials if there is ever a conflict with a pre-existing value.
This column may not be empty, it must contain characters, and it must contain at least one non-whitespace character.
The initials, or notes regarding the initials, used to identify the person when recording GPS data.
Note
This column exists because of a historical inconsistency between the initials used in the collection of GPS data and the initials used in the collection of other data. It is strongly recommended that new observers use the same initials when collecting either sort of data.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The person's real name.
This column may not be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
Textual remarks regarding when the observer was recording Babase data. Usually a date range.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
Any notes you may wish to make on the person. For example you may wish to record that the person is John Smith the graduate student, not John Smith the, for example, President of Kenya who asked to be able to collect data and actually did so for a day.
This column may be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The role the person has filled in regards to data collection. Must be a value on the OBSERVER_ROLES table.
This column must not be NULL.
The SWERB_OBSERVERS.Role value to use when the observer is identified in the first line of SWERB data supplied to the SWERB_UPLOAD view. Must be a value on the OBSERVER_ROLES table.
This column must not be NULL.
The SWERB_OBSERVERS.Role value to use when the observer is identified in the second line of SWERB data supplied to the SWERB_UPLOAD view. Must be a value on the OBSERVER_ROLES table.
This column must not be NULL.
One row for every role a person may have in data collection.
OBSERVER_ROLES defines vocabulary for OBSERVERS.Role and SWERB_OBSERVERS.Role.
The different reasons why a focal individual's neighbor is unable to be identified during focal point sampling or the code(s) used to identify lone males when recording SWERB other group observations.
A Unksname value must not appear as a BIOGRAPH.Sname value.
UNKSNAMES defines vocabulary for the Unksname column of the NEIGHBORS table. It is also used by the SWERB_UPLOAD view to identify unknown lone males.
A boolean. When TRUE the Unksname value is used to indicate
that an unknown lone male was observed during a SWERB other
group observation; FALSE otherwise.
This column may not be NULL.
The categories of accuracy of the birth date estimates.
BSTATUSES defines values for the Bstatus column of BIOGRAPH. Except for the "unknown" Bstatus
(9.0), this column indicates
the length in years of the estimated range of an
individual's possible birth dates.
For example, a Bstatus
1 indicates that the individual is
estimated to have been born within 1
year of the Birth date, or the
Birth date plus or minus at most
6 months.
The value 9.0
(unknown) has a special meaning to the system. This is the
only BIOGRAPH.Bstatus value that indicates that the
individual's birth date is "unknown", i.e. not able to be
estimated with any meaningful confidence. It is the only
Bstatus value that allows an
individual to have a NULL EarliestBirth and LatestBirth.
The value 9.0
(unknown) is also the only Bstatus value whose numeric value has
no actual meaning as a number. All
other numbers added to BSTATUSES are
presumed to indicate a number of years of accuracy in a
birth date estimate.
The possible degrees of confidence in the nature and agent of the recorded cause of death (the recorded BIOGRAPH.Dcause values), recorded disperse date (the recorded DISPERSEDATES.Dispersed), and recorded maternal group assignment (the recorded BIOGRAPH.Matgrp) values.
The values in this table are used to indicate confidence in several different tables, so it is necessary to describe these values in general terms. Unfortunately, this intentional lack of specificity in description may cause an unintentional lack of clarity. For this reason, the textual column Usage is included, in which table-specific comments or clarifications may be added.
CONFIDENCES defines values for the DcauseNatureConfidence and DcauseAgentConfidence columns of the BIOGRAPH table, the Dispconfidence column of the DISPERSEDATES table, and the Matgrpconfidence column of the BIOGRAPH table.
The value 0
(not applicable) has a special meaning to the system. This
is the only BIOGRAPH.DcauseNatureConfidence and DcauseAgentConfidence value allowed to be
associated with individuals having no cause of death, having
a Dcause of
0.
This support table indicates the apparent completeness of a paternity assignment.
DADS_COMPLETENESS defines values for the Completeness column of DADS_CONSENSUS.
The possible kinds of data from paternity analyses.
DADS_EVIDENCE_TERMS defines values for the Term column of DADS_EVIDENCE.
This column indicates what kind of data is expected to be recorded as a Datum with this Term. Its legal values are:
BOOLThis Data_Type indicates that the Datum is expected to be a boolean, either
TRUEorFALSE.DATEThis Data_Type indicates that the Datum is expected to be a date, recorded in YYYY-MM-DD format.
NUMThis Data_Type indicates that the Datum is expected to be a number, e.g.
12or12.0but nottwelve.TEXTThis Data_Type indicates that the Datum is expected to be plain text, i.e. it has no restrictions.
It is an error if the Datum value does not match the
format of its Term's
Data_Type. For example, if the Term PCR
DATE has a data type of
DATE, then a DADS_EVIDENCE row will return an error if its
Datum is anything other than a date in YYYY-MM-DD
format.
This column may not be NULL.
This boolean column indicates whether this row's Term
is unique per analysis. When TRUE, this Term cannot be
used in DADS_EVIDENCE more than once per
analysis.
This column cannot be NULL and is TRUE by
default.
This support table categorizes the type(s) of genetic mismatches that are observed in a paternity assignment.
DADS_MISMATCHES defines values for the Mismatch column of DADS_CONSENSUS.
The different causes of death, classified by Nature and Agent.
A Dcause's Nature describes in general terms the reason that the individual died, e.g. “violence”. A Dcause's Agent indicates the source or cause of the indicated nature, e.g. “predator”.
When an individual's Dcause is assigned, the assignment is often not certain, but is instead a CONFIDENCES-qualified inference based on available evidence at the time of the individual's death or disappearance. Because of this, an individual's Dcause should be read as "if Nature, then Agent".
The combination of Nature and Agent is unique.
See the note in the STATUSES: Special Values section for an explanation of what it means to be alive.[229]
The value 0 (no cause of death) has a special meaning to the system. This is the only Dcause allowed to be associated with living individuals.
Indicates the data source of demography notes.
DEMOG_REFERENCES defines values for the Reference column of DEMOG.
The different natures, or reasons, for death. See DCAUSES for more info about the difference between causes, natures, and agents of death.
DEATHNATURES defines values for the Nature column of DCAUSES.
Indicates the different ways individuals enter the study population.
ENTRYTYPES defines values for the Entrytype column of BIOGRAPH.
A boolean that indicates if individuals with this Entrytype should automatically be assigned residency on their Entrydate. See Entrydate is Special for more information.
The value of B
(birth) has a special meaning to the system. When an
individual has this Entrytype,
their BIOGRAPH.Entrydate must also be the date of
their Birth. In the residency rules,
individuals with Entrytype
B are assigned a LowFrequency of FALSE on their Entrydate and subsequent
28 days, regardless of the actual
number of censuses that occurred during that period. No
other value should be used in BIOGRAPH.Entrytype to
indicate birth as the method for entry into the
population.
The Residency_Special_Case for Entrytype
B must be
TRUE.
The different meanings of various maturity marker date values.
MSTATUSES defines values for MATUREDATES.Matured and RANKDATES.Ranked columns.
Tip
May be O (ON) or
B
(BY). O indicates a known
date. B indicates that we know
that the animal had reached that maturational marker BY
the given date but we have no information about the actual
date on which the marker was attained.
The different categories of rankings that order individuals by dominance within a group within a month. Each category of ranking is identified with a row of this table.
This table contains a “special” column, Query. The Query column is an SQL query which defines which individuals are eligible for inclusion in this category of ranking. The SQL statement determines which individuals are included in any given ranking. It must return distinct Snames of individuals to be ranked within a given group over a given time period. In general the query is a SELECT statement which uses the BIOGRAPH and MEMBERS tables to determine who is to be ranked within a group over a month. A number of “special symbols” may be, and will need to be, included in the SQL query. Each “special symbol” represents a value which changes depending on the month or group ranked. The “special symbols” are:
| Notation | Mnemonic | Data type | Description of usage |
|---|---|---|---|
| %g | Group id | number | The Gid of the group being ranked. |
| %s | Start date | date | (Should not be quoted in the SQL statement.) Date of the first day of the interval over which the individuals are ranked (inclusive.) |
| %f | Finish date | date | (Should not be bracketed in the SQL statement. Date of the last day of the interval over which the individuals are ranked (inclusive.) Note that ages, maturation dates, and so forth are often computed using or compared to the Finish Date value. |
The different states of an individual, reflecting what sort of record keeping needs to be done on the individual in the future.
A boolean that indicates if individuals with this Status should be able to retain residency through their Statdate when terminal absences might suggest otherwise. See Statdate is (also) special for more information.
The value 0 (alive) has a
special meaning to the system. No other codes should be
created to indicate that the individual is alive.
Note
“Alive” has a particular meaning to Babase. It does not mean “alive in real life”, a concept which itself is complicated when it is qualified by a time because there are not always recorded observations.
Alive in the context of Babase means “an individual on which data is continuing to be collected”. The foremost implication of this is that living individuals can have data added to Babase after the individuals' BIOGRAPH.Statdate and the BIOGRAPH.Statdates will automatically change. Babase will hold no data on an individual that postdates the individual's death. The only way the BIOGRAPH.Statdate of a dead individual changes is when the change is made manually.
The different hormones that may be extracted and analyzed.
HORMONE_IDS defines values for the Hormone column of HORMONE_KITS.
The code used to uniquely identify the hormone.
This column may not be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The value E has a
special meaning to the system. This is the only HORMONE_KITS.Hormone value that indicates the
kit measures concentration of estrogen. Also, the value is
used in the ESTROGENS view.
The value GC has a
special meaning to the system. This is the only HORMONE_KITS.Hormone value that indicates the
kit measures concentration of glucocorticoids. Also, the
value is used in the GLUCOCORTICOIDS
view.
The value P has a
special meaning to the system. This is the only HORMONE_KITS.Hormone value that indicates the
kit measures concentration of progesterone. Also, the value
is used in the PROGESTERONES view.
The value T has a
special meaning to the system. This is the only HORMONE_KITS.Hormone value that indicates the
kit measures concentration of testosterone. Also, the value
is used in the TESTOSTERONES view.
The value TH has
a special meaning to the system. This is the only HORMONE_KITS.Hormone value that indicates the
kit measures concentration of thyroid hormone. Also, the
value is used in the THYROID_HORMONES
view.
The different procedures that may be performed in preparation for hormone analyses.
HORMONE_PREP_PROCEDURES defines values for the Procedure column of HORMONE_PREP_DATA.
The code used to uniquely identify the procedure.
This column may not be NULL. This column may not be empty, it must contain characters,
and it must contain at least one non-whitespace character.
The value MEOH_EXT has
a special meaning to the system. This is the only HORMONE_PREP_DATA.Procedure value that indicates
a methanol extraction. Also, this value is used in the
ESTROGENS, GLUCOCORTICOIDS, PROGESTERONES, and TESTOSTERONES views.
The value SPE has a
special meaning to the system. This is the only HORMONE_PREP_DATA.Procedure value that indicates
a solid-phase extraction. Also, this value is used in the
ESTROGENS, GLUCOCORTICOIDS, PROGESTERONES, and TESTOSTERONES views.
The value ETOH_EXT has
a special meaning to the system. This is the only HORMONE_PREP_DATA.Procedure value that indicates
an ethanol extraction. Also, this value is used in the
THYROID_HORMONES view.
The different software types used for genetic hybrid score analyses.
HYBRIDGENE_SOFTWARE defines values for the Software column of HYBRIDGENE_ANALYSES.
The genetic marker types used in genetic hybrid score analyses.
MARKERS defines values for the Marker column of HYBRIDGENE_ANALYSES.
The different categories of healing progress used in wound/pathology healing updates.
WP_HEALSTATUSES defines values for the HealStatus column of WP_HEALUPDATES.
The possible statuses describing the current state of a wound/pathology report.
WP_REPORTSTATES defines values for the ReportState column of WP_REPORTS.
The codes defining all the possible wounds and pathologies.
The ImpairsLocomotion and InfectionSigns columns affect
validation of the identically-named columns in WP_DETAILS. For each code, all WP_DETAILS.ImpairsLocomotion values must equal the
related ImpairsLocomotion
in this table, unless this table's ImpairsLocomotion is
NULL. Likewise with InfectionSigns, for each code, all
WP_DETAILS.InfectionSigns values must equal the
related InfectionSigns in
this table, unless this table's InfectionSigns is NULL.
WP_WOUNDPATHCODES defines values for the WoundPathCode column of WP_DETAILS.
A character, indicating the required WP_DETAILS.ImpairsLocomotion value, if any, for this WoundPathCode.
This column may be NULL, when there is no required
ImpairsLocomotion for this
WoundPathCode.
A character, indicating the required WP_DETAILS.InfectionSigns value, if any, for this WoundPathCode.
This column may be NULL, when there is no required
InfectionSigns for this
WoundPathCode.
The activities recorded in focal point observations.
ACTIVITIES defines values for the Activity of the POINT_DATA table and the Activity of the STYPES_ACTIVITIES table.
The value F has a
special meaning to the system. It indicates feeding and
triggers a required Foodcode in
POINT_DATA.Foodcode.
The different kinds of (non-multiparty) interactions between individuals which may be recorded.
The various kinds of interactions may be grouped together into larger categories, which are themselves valid kinds of interactions. The Class column is used for this purpose. The Class column contains an Act value identifying the larger class of interactions to which the interaction belongs. If the interaction does not belong to a larger category, the Class should contain the row's own Act value. Only 1 level of classification hierarchy is allowed -- the ACTS row referenced in the Class column must have a Class value equal to its Act value.
Rows that contain a TRUE value in the Retired column
may not be referred to by newly created database rows,
although, presumably older, pre-existing rows may contain the
Act values of these retired rows. Should it be necessary to
create such new rows, retired ACTS may be temporarily
un-retired.
ACTS defines values for the Act column of INTERACT_DATA.
All of the Class codes on ACTS have a special meaning to the system's programs. New Class codes may not be created and rows that represent the classifications, have an Act equal to their Class, cannot have their Act, Class, or Descr values changed.[230]
The ACTS row of interaction classification. (See
above.) This column may not be NULL.
One row for each version of data structure produced by Psion devices when exporting focal sampling data.
Note
The primary purpose of this table is to ensure that the data coming off a Psion unit is correctly interpreted by the Psionload program and loaded into the right tables. The structure and semantics of data collected by a Psion unit is determined by the setup file, but various setup files can produce the same output.
See below for information about the relevance of these values to focal data that did not come from a Psion device.
For further comments, see SETUPIDS.
DATA_STRUCTURES defines vocabulary for SETUPIDS.Data_Structure.
The Data_Structure value
1 is the data
structure understood by the Psionload
program.
The social contexts in which multiparty interactions occur.
CONTEXT_TYPES defines values for MPIS.Context_type.
Special CONTEXT_TYPES
NNo context. The MPIS.MPIS-Context column must be
NULLwhen this code is used.CConsortship. The multiparty interaction occurred in the context of a consortship.
The MPIS.MPIS-Context column must be
NULLwhen this code is used. The MPIS.Context_type column must beCwhen a related CONSORTS row exists. The system will generate a warning when the MPIS.Context_type column isCand there is no related CONSORTS row.
The different food items eaten by baboons.
FOODCODES defines values for POINT_DATA.Foodcode.
Food items are themselves categorized into types. This column contains the type of the food item. Valid food type values are those stored in the FOODTYPES.Ftype column.
There are no special FOODCODE values, however it is
worth remarking on the POINT_DATA.Activity
F value, which has
special meaning to the system. The POINT_DATA.Foodcode
column must contain a value when and only when POINT_DATA.Activity
is F, otherwise POINT_DATA.Foodcode
must be NULL.
Food items are categorized into broader classifications using the codes defined on the FOODTYPES tables.
The different spatial relationships between mother and infant recorded during adult female all-occurrences point sampling.
KIDCONTACTS defines vocabulary for the Kidcontact column of the FPOINTS table.
The different kinds of dyadic interactions which may be recorded as interactions occurring during a multiparty interaction event. There are 4 mutually exclusive categories of interactions: Agnoisims, Requests for help, Help given, and Other.
The Decided column cannot be TRUE unless the kind of
the act is an agonism -- the Kind column is
A.
Note
Although Babase stores multiparty interactions using a data structure similar to that used to store non-multiparty interactions the data sets are separate, different kinds of interactions are recorded using different codes, and the interactions are never recorded in both data sets.
The value AH must be
the code used to indicate the giving of active help. The
value PH must be the code
used to indicate the giving of passive help. These codes
are tested for in the process of generating warnings
indicating that the MPI_DATA.Active value may be incorrect.
Some values have special meaning to the MPI_UPLOAD view, in that the view changes act values in the uploaded file to particular values. See the documentation on this for more detail.
This column classifies the kind of interaction into one of 4 distinct types, as listed below.
MPIACTS.Kind values
AAn Agonism interaction.
RA Request for help.
HHelp given.
OOther.
This column may not be NULL.
The different classifications of neighbor recorded during focal point observations.
The Requires, Nsex, and Nunique columns allow for some more complicated validation of Ncode use in NEIGHBORS, as discussed below.
When neighbors should be recorded in a specific order,
the Requires column ensures that they
are. When there is a value in this column, the row's Ncode
cannot be used as a NEIGHBORS.Ncode for the point observation (the
NEIGHBORS.Pntid)
unless that point already has another NEIGHBORS row with this NCODES row's Requires. For example, suppose Ncode
2 indicates the second nearest neighbor and
Ncode 1 is the nearest neighbor[231]. When Ncode 1 is placed in
Ncode 2's Requires column, Babase will not
allow a point observation to have a second nearest neighbor
(Ncode 2) unless there is already a nearest
neighbor (Ncode 1).
The Nsex column is used to enforce that a neighbor must be a particular sex. This is complicated because it may rely on the sex of the neighbor with the Ncode specified in the Requires column, as discussed below.
An NCODES row may not have a Requires of NULL and a Nsex value of
O.
In some sampling protocols, one individual might be the
appropriate Sname for more than
one Ncode. In other cases, it may be preferable to enforce
that all neighbors recorded in a point observation be
distinct. This type of validation is controlled by the
boolean Nunique column. When TRUE,
the Sname must be unique among all
the neighbors of a particular point observation (NEIGHBORS.Pntid).
When FALSE, the Sname need not
be unique.
NCODES defines vocabulary for the Ncode column of the NEIGHBORS table and the Ncode of the STYPES_NCODES table.
Another Ncode, representing the neighbor type that must be recorded in a point observation before this row's Ncode can be recorded with that observation.
This column may be NULL, in which case the only
requirement is that the same Ncode not be used twice in one
point observation.
The sex that the neighbor must have. Possible values and their meanings:
| Code | Mnemonic | Definition |
|---|---|---|
| A | Any | The neighbor with this Ncode may be of any sex. |
| M | Male | The neighbor with this Ncode must be male.[232] |
| O | Opposite | The neighbor with this Ncode must be of a different sex than the neighbor with the Requires Ncode. Note that because there are 3 sexes — male, female, and unknown — this does not strictly conform with the field monitoring guide which only takes males and females into account. If this is a problem then we need to do something about it. |
Caution
Neighbors with a Unksname rather than a Sname are always considered to be of
the opposite sex — they satisfy the
O Nsex code.
This column may not be NULL.
The different reasons why a participant in a multiparty interaction is unable to be identified during data collection.
A Unksname value must not appear as a BIOGRAPH.Sname value.
PARTUNKS defines vocabulary for the Unksname column of the MPI_PARTS table. It is also used by the MPI_UPLOAD view to test the uploaded data for unknown participants in a consortship interaction.
The postures recorded in focal point observation.
POSTURES defines values for POINT_DATA.Posture and STYPES_POSTURES.Posture.
One row for each version of each program used on a handheld data collection device.
Note
The primary purpose of this table is to avoid storing relatively lengthy identical strings on the SAMPLES table. This table would probably not be worth having were not the program ID strings reported by the devices so long, and did we not need the SETUPIDS table, which is very similar to this table.
One row for each device or "system" used for collecting focal sample data.
Note
Originally, this table explicity listed only Psion focal sampling units and was named “PALMTOPS”. (At the time, that name was presumed to be an appropriately generic description for any kind of mobile electronic device.) In Babase 5.5.2, the table was renamed because 1) the term “palmtop” comes from another era and has little to no meaning for modern users, and more importantly 2) in preparation for the expected addition of focal data that were collected with only a pen and paper, the name needed to be changed to be more inclusive anyway. Ideally, the PALMTOPS_HISTORY table should have remained in the babase_history schema so that changes to it would remain accessible. However, when this table was renamed the PALMTOPS_HISTORY table was empty. There were no archived changes that needed to be preserved, so the PALMTOPS_HISTORY table was not retained.
SAMPLES_COLLECTION_SYSTEMS defines vocabulary for SAMPLES.Collection_System.
One row for each configuration — which may represent one or more specific files — used in a program for data collection.
Note
Although not every setup file can be used with every version of every program, Babase makes no attempt to validate the setup files against the program files, or vice versa. This is because the data are expected to be generated by the programs and, unless they lie about the program they are running and the setup file used, whatever program id is reported must, ipso facto, work with the reported setup file.
Note
The primary purpose of this table is to ensure, via its relation with the DATA_STRUCTURES table, that the data coming off the device is correctly interpreted by the Psionload program and loaded into the right tables. The table also allows Babase to save space on the SAMPLES table by storing the small Setupid integer rather than the relatively long setup ID strings reported by the devices.
Note
The Data_Structure column is only used by the Psionload program. When a Setupid appears in SAMPLES with a Collection_System that is not a Psion device, its data are not expected to be imported via the Psionload program so the Setupid's Data_Structure value is irrelevant.
The system makes no attempt to validate the Data_Structure against the Collection_System for the reasons discussed above, not to mention that Psion units and the Psionload program are legacy systems with no modern use.
Warning
The setupid should determine the structure and semantics of the device's data files. If this assumption is violated, e.g. by having two different Psion programs produce different results from the same setup file, then the Psionload program may do bad things to the database.
For further comments, see PROGRAMIDS.
The DATA_STRUCTURES.Data_Structure indicating the version of the data structure produced by devices using the setup file.
This column may not be NULL.
The different focal sampling protocols used, including several columns that indicate how a sample's data should be validated.
The Sex column indicates if
this row's sampling protocol is specific to individuals of a
particular Sex. When this column
is not NULL, SAMPLES rows with this SType must have an Sname of an individual whose BIOGRAPH.Sex matches
this row's Sex.
The Max_Points column indicates the maximum number of points that are allowed to be recorded for a sample with this SType. All SAMPLES rows with this SType must have Mins and Minsis values less than or equal to this value.
The Has_FPoints column
indicates if points from samples with this SType are allowed
to include data about the focal individual's infant. When
TRUE, a SAMPLES row with this SType can have its related Pntid's in FPOINTS.
Many focal sampling protocols are explicitly targeted toward individuals of a specific age class, e.g. "adults" or "juveniles". Validating that an individual is in a particular age/sex class usually involves comparing the SAMPLES.Date to a certain "milestone" date in the individual's life, e.g. their MATUREDATES.Matured or RANKDATES.Ranked. For various reasons it is often desirable to allow some "wiggle room" when using these dates. For example, if males are considered "adults" on or after their RANKDATES.Ranked then a rule could be made requiring that samples on "adult males" must never be before the focal individual's Ranked date, but instead it may be preferable to allow samples on "adult males" to be some small period of time before his Ranked date. This table includes several columns that enable that kind of validation.
For each "milestone" date of interest, there is a Days_Before_Xxx column, a Days_After_Xxx column, and a Req_Xxx column. Validation related to the MATUREDATES.Matured is enabled using the Days_Before_Matured, Days_After_Matured, and Req_Matured columns; validation related to the RANKDATES.Ranked is enabled using the Days_Before_Ranked, Days_After_Ranked, and Req_Ranked columns; and validation related to the BIOGRAPH.Birth date of a female's first offspring is enabled using the Days_Before_FirstBirth, Days_After_FirstBirth, and Req_FirstBirth columns.
A Days_Before_Xxx column contains an integer that
indicates the maximum number of days before the Xxx date on
which a focal sample may occur with the indicated SType. E.g.
a row's Days_Before_Matured is some
number n, indicating that the Date of all SAMPLES rows
with this row's SType cannot be more
than n days before the focal individual's
Matured date[233].
A Days_After_Xxx column contains an integer that
indicates the maximum number of days after the Xxx date on
which a focal sample may occur with the indicated SType. E.g.
a row's Days_After_Matured is some
number n, indicating that the Date of all SAMPLES rows
with this row's SType cannot be more
than n days after the focal individual's
Matured date[234].
In many cases, the individual will not have a
"milestone" date in the database for legitimate
reasons[235]. Because of this, the Days_Before_Xxx and
Days_After_Xxx columns will not provoke an error when the
focal individual does not have an Xxx date. However, for some
sampling protocols it may be desirable to
require that the focal individual have an
Xxx date. This requirement can be toggled via the Req_Xxx
column. E.g. when the Req_Matured
column is TRUE, a SAMPLES row with this
SType must have an Sname that appears in the MATUREDATEStable.
Presumably, a focal sampling protocol that requires a
certain "milestone" date will likely also have some rules
using that date to validate the sample's Date. The system will return a warning
for any STYPES rows with a TRUE Req_Xxx but whose related
Days_Before_Xxx and Days_After_Xxx columns are NULL.
Some focal sample types can be described as following a "representative interactions" protocol, which involves observers collecting all interactions in their line of sight in the course of collecting focal samples on all individuals of a given age-sex class in a random order. The Repr_Interxns column indicates which sample types follow that protocol. See the discussion with the ACTOR_ACTEES view for more information about the utility of this designation.
STYPES defines the vocabulary for the SAMPLES.SType, STYPES_ACTIVITIES.SType, STYPES_POSTURES.SType, and STYPES_NCODES.SType columns.
The required BIOGRAPH.Sex of all focal individuals with this SType.
This column may be NULL, indicating that this SType
does not require that the focal individual be a specific
sex.
The maximum allowed number of points in a focal sample of this SType.
This column must be a positive integer and cannot be
NULL.
A boolean indicating if focal samples of this SType can have related rows in FPOINTS.
This column may not be NULL.
A non-negative integer, indicating the largest number of days before the focal individual's MATUREDATES.Matured (if any) on which focal samples of this SType are allowed.
This column may be NULL, indicating that samples
with this SType can be any number of days before the focal
individual's Matured
date.
A non-negative integer, indicating the largest number of days after the focal individual's MATUREDATES.Matured (if any) on which focal samples of this SType are allowed.
This column may be NULL, indicating that samples
with this SType can be any number of days after the focal
individual's Matured
date.
A boolean, indicating whether focal individuals in samples of this SType are required to have a MATUREDATES.Matured date.
This column may not be NULL.
A non-negative integer, indicating the largest number of days before the focal individual's RANKDATES.Ranked (if any) on which focal samples of this SType are allowed.
This column may be NULL, indicating that samples
with this SType can be any number of days before the focal
individual's Ranked date.
A non-negative integer, indicating the largest number of days after the focal individual's RANKDATES.Ranked (if any) on which focal samples of this SType are allowed.
This column may be NULL, indicating that samples
with this SType can be any number of days after the focal
individual's Ranked date.
A boolean, indicating whether focal individuals in samples of this SType are required to have a RANKDATES.Ranked date.
This column may not be NULL.
A non-negative integer, indicating the largest number of days before the BIOGRAPH.Birth of the focal individual's first offspring (if any) on which focal samples of this SType are allowed.
This column may be NULL, indicating that samples
with this SType can be any number of days before the Birth of the focal individual's first
offspring.
A non-negative integer, indicating the largest number of days after the BIOGRAPH.Birth of the focal individual's first offspring (if any) on which focal samples of this SType are allowed.
This column may be NULL, indicating that samples
with this SType can be any number of days after the Birth of the focal individual's first
offspring.
Vocabulary describing which Activity values are allowed to be used with each SType. There is one row for each Activity allowed to be used with each SType.
Unlike most other support tables, this table does not have a Descr column. This table exists only to define vocabulary, so the meaning or "description" of each row is fully explained by the values in the SType and Activity columns.
Each SType-Activity dyad must be unique.
Tip
This table is important for data management but has little or no practical use for regular database users. If you are not a data manager, it is probably safe for you to ignore this table altogether.
STYPES_ACTIVITIES defines the vocabulary for the use of two separate columns in separate but related tables: SAMPLES.SType and POINT_DATA.Activity.
Each of those columns has its own support table controlling the vocabulary for its respective column (SType has STYPES, Activity has ACTIVITIES), but this table defines how those columns' values are allowed to be used together.
This table does not use a single column as its key. Instead, the key is the combination of SType and Activity. Those columns' separate values are the ones used in SAMPLES and POINT_DATA, so there is no utility gained from creating a separate "key" column here.
The STYPES.SType of the sample type in which this row's Activity is allowed to be used.
This column may not be NULL.
The ACTIVITIES.Activity that is allowed to be used with this row's SType.
This column may not be NULL.
Vocabulary describing which Ncodes are allowed to be used with each SType. There is one row for each Ncode allowed to be used with each SType.
Unlike most other support tables, this table does not have a Descr column. This table exists only to define vocabulary, so the meaning or "description" of each row is fully explained by the values in the SType and Ncode columns.
Each SType-Ncode dyad must be unique.
Tip
This table is important for data management but has little or no practical use for regular database users. If you are not a data manager, it is probably safe for you to ignore this table altogether.
STYPES_NCODES defines the vocabulary for the use of two separate columns in separate but related tables: SAMPLES.SType and NEIGHBORS.Ncode.
Each of those columns has its own support table controlling the vocabulary for its respective column (SType has STYPES, Ncode has NCODES), but this table defines how those columns' values are allowed to be used together.
This table does not use a single column as its key. Instead, the key is the combination of SType and Ncode. Those columns' separate values are the ones used in SAMPLES and NEIGHBORS, so there is no utility gained from creating a separate "key" column here.
Vocabulary describing which Postures are allowed to be used with each SType. There is one row for each Posture allowed to be used with each SType.
Unlike most other support tables, this table does not have a Descr column. This table exists only to define vocabulary, so the meaning or "description" of each row is fully explained by the values in the SType and Posture columns.
Each SType-Posture dyad must be unique.
Tip
This table is important for data management but has little or no practical use for regular database users. If you are not a data manager, it is probably safe for you to ignore this table altogether.
STYPES_POSTURES defines the vocabulary for the use of two separate columns in separate but related tables: SAMPLES.SType and POINT_DATA.Posture.
Each of those columns has its own support table controlling the vocabulary for its respective column (SType has STYPES, Posture has POSTURES), but this table defines how those columns' values are allowed to be used together.
This table does not use a single column as its key. Instead, the key is the combination of SType and Posture. Those columns' separate values are the ones used in SAMPLES and POINT_DATA, so there is no utility gained from creating a separate "key" column here.
The STYPES.SType of the sample type in which this row's Posture is allowed to be used.
This column may not be NULL.
The POINT_DATA.Posture of the Posture that is allowed to be used with this row's SType.
This column may not be NULL.
Classifies samples collected during a darting into specific categories, e.g. blood, skin, etc.
DART_SAMPLE_CATS defines values for DART_SAMPLE_TYPES.DS_Cat. This column cannot be changed.
The different types of samples that are collected during dartings.
This table contains data that are special values used by the DSAMPLES view. Because of this, only administrators are allowed to INSERT, UPDATE, or DELETE from this table.
DART_SAMPLE_TYPES defines values for DART_SAMPLES.DS_Type. This column cannot be changed.
The values in DS_Type are used in the definition of the DSAMPLES view.
The DART_SAMPLE_CATS.DS_Cat to which each sample type belongs.
Some sample types may be sex-specific. Vaginal and cervical swabs, for example, can only be collected from females. If a sample type has any such specificity, the correct sex is indicated here.
This column may be NULL when the darting sample is
not sex-specific.
The minimum number of samples of this type that can be
collected. This column may not be NULL.
The different anesthetics used when darting.
The different conditions a lymph node can be found in when darting.
The different kinds of parasites, kinds of parasites in varying developmental stages, or kinds of parasite indicators counted when darting.
The different tooth conditions, degrees of wear, chipping, etc., observed when darting.
Caution
The condition of the tooth is a property distinct from the degree to which the tooth is present or absent. The latter property is described by the codes in the TSTATES table.
TCONDITIONS defines values for TEETH.Tcondition.
The classifications of parasite count useful in analysis.
TICKSTATUSES defines values for TICKS.Tickstatus.
The following special codes can only be altered by suitably privileged individuals. See Special Values.
Special TICKSTATUSES codes
0A count was performed and no parasites were found. This code can only be used when the number of parasites counted is
0, in which case it must be used.1A count was performed and parasites were found. This code must be used when the number of parasites counted is any non-zero positive integer. The code may be used when parasites are found but were not counted (TICKS.Tickcount is
NULL).
A set of codes describing the dentition of a baboon, one code for each tooth.
Note
Deciduous[236] teeth have different codes than, are considered different from, adult teeth.
Every toothcode value is special, although there are no restrictions placed upon making changes to these “special” values as there are on the special rows in other tables. Each of the TOOTHCODES.Tooth values are written into[237]the DENT_CODES and DENT_SITES views. Adding or deleting rows from TOOTHCODES requires re-writing the DENT_CODES and DENT_SITES views to ensure the alterations are present in the views.
Boolean value indicating whether the tooth is a canine or not.
Note
Morphologically this column should be on TOOTHSITES, be associated with tooth location. Placement on this table allows control over whether canine data may be collected on decidious teeth -- control which is not needed at this time.
This column may not be NULL.
Boolean value indicating whether the tooth is
deciduous or adult. TRUE indicates the tooth is
deciduous. FALSE indicates the tooth is adult.
This column may not be NULL.
I am inclined to make the name of this column be Adult rather than Deciduous for reasons of brevity, but I believe that Susan prefers it as-is. I would like feedback from the folks who are likely to be doing the typing. (KOP)
The site of the tooth within the mouth. Legal values for this column are defined by the TOOTHSITES table.
This column may be used to correlate the locations of deciduous teeth with their adult counterparts.
This column may not be NULL.
I am inclined to make the name of this column be Site, but I believe that Susan prefers it as-is. I would like feedback from the folks who are likely to be doing the typing. (KOP)
The locations of a baboon's teeth within the mouth. This table is used to correlate adult with deciduous teeth. Any given TOOTHSITES code cannot be used in two TOOTHCODES rows having the same TOOTHCODES.Deciduous value -- at most one adult and one deciduous tooth can have the same location within the mouth.
TOOTHSITES defines values for TOOTHCODES.Toothsite.
Codes describing the degree to which a tooth is present or absent in the mouth.
Caution
The degree to which the tooth is present or absent is a property distinct from the condition of the tooth. The latter property is described by the codes in the TCONDITIONS table.
The value M
(missing) has a special meaning to the system. TEETH rows that describe missing teeth must have
NULL TEETH.Tcondition values.
The possible locales where tissue and nucleic acid samples can be stored or used.
INSTITUTIONS defines values for the Institution column in LOCATIONS, NUCACID_DATA, NUCACID_LOCAL_IDS, TISSUE_DATA, and TISSUE_LOCAL_IDS.
The value 1 has
special meaning to the system. It is used in the TISSUES, NUCACIDS, and NUCACIDS_W_CONC views to help populate each
view's respective LocalId_1 column.
The value 2 has
special meaning to the system. It is used in the TISSUES, NUCACIDS, and NUCACIDS_W_CONC views to help populate each
view's respective LocalId_2 column.
The possible levels of confidence in the identity of a tissue sample.
MISID_STATUSES defines values for TISSUE_DATA.Misid_Status.
The possible methods for quantifying nucleic acid concentrations.
NUCACID_CONC_METHODS defines values for NUCACID_CONC_DATA.Conc_Method.
The value 1 has a
special meaning to the system. This is the only NUCACID_CONC_DATA.Conc_Method value that
indicates quantification by quantitative PCR ("qPCR"). Also,
this value is used in the definition of
the NUCACIDS_W_CONC view.
The value 2
has a special meaning to the system. This is the only NUCACID_CONC_DATA.Conc_Method value that
indicates quantification with a Nanodrop
spectrophotometer. Also, this value is used in the definition of
the NUCACIDS_W_CONC view.
The value 3 has
a special meaning to the system. This is the only NUCACID_CONC_DATA.Conc_Method value that
indicates quantification with a Qubit fluorometer. Also,
this value is used in the definition of
the NUCACIDS_W_CONC view.
The value
4 has a special
meaning to the system. This is the only NUCACID_CONC_DATA.Conc_Method value that
indicates quantification with a Bioanalyzer assay. Also,
this value is used in the definition of
the NUCACIDS_W_CONC view.
The value 5
has a special meaning to the system. This is the only NUCACID_CONC_DATA.Conc_Method value that
indicates quantification with a Quant-iT assay. Also, this
value is used in the definition of
the NUCACIDS_W_CONC view.
The possible units used in quantifications of nucleic acid concentrations.
The values in this table are used by the convert_conc() to convert values from one unit to
another. This is possible only when both units are related.
The Reference column is
used to indicate that relatedness. It is a Unit value that is used as a
standard or point of reference for all the units in that
group. E.g. rows for the unit μM
(micromolar) and nM (nanomolar) might both
have a Reference of M (molar)[238].
The Conversion
column is a number that indicates how the Unit is related to
its Reference. Given a quantity whose unit is the Reference,
multiply that quantity by the Conversion to get a
concentration with that row's Unit. E.g. in a row with Unit
mM (millimolar) and Reference
M (molar), the Conversion should be
1000.
When the Unit and
Reference are equal, the
Conversion must be
1. When Unit and Reference are not equal,
the Conversion cannot be 1.
NUCACID_CONC_UNITS defines values for NUCACID_CONC_DATA.Unit and for the two "unit" parameters of the convert_conc() function.
The value NG/UL has a
special meaning to the system. This is the only NUCACID_CONC_DATA.Unit value that indicates
nanograms per microliter (ng/μL). Also, this value is
used in the NUCACIDS_W_CONC view to show
several concentrations in ng/μL.
The value PG/UL has a
special meaning to the system. This is the only NUCACID_CONC_DATA.Unit value that indicates
picograms per microliter (pg/μL). Also, this value is
used in the NUCACIDS_W_CONC view to show
a concentration in pg/μL.
The Unit used for reference when converting quantities to and from this row's Unit.
This column may not be NULL.
The possible methods for creating nucleic acid samples.
NUCACID_CREATION_METHODS defines values for NUCACID_DATA.Creation_Method.
The possible nucleic acid sample types.
NUCACID_TYPES defines values for NUCACID_DATA.NucAcid_Type.
This column may not be empty, it must contain characters, and it must contain at least one non-whitespace character.
The possible media used for storage/archiving of tissue samples.
STORAGE_MEDIA defines values for TISSUE_DATA.Storage_Medium.
This column may not be empty, it must contain characters, and it must contain at least one non-whitespace character.
The possible tissue sample types.
Some types of tissues, e.g. blood, cannot plausibly be collected after an individual has died or disappeared. Other tissue types may be collected during regular observation or sometime afterward, e.g. "skin" could be a puncture from a live animal or a patch of dry flesh from a found corpse. To validate the TISSUE_DATA.Collection_Date against the source individual's BIOGRAPH.Statdate, but also allow the flexibility to set different rules for different tissue types, this table includes the Max_After_Statdate column.
When the Max_After_Statdate column is not
NULL, all TISSUE_DATA rows that have this
row's Tissue_Type and that came from an individual in the main
population cannot have a Collection_Date that is more than
Max_After_Statdate days after
the individual's Statdate. That is,
tissue samples cannot be collected more than Max_After_Statdate days after the
individual's death/disappearance.
TISSUE_TYPES defines values for TISSUE_DATA.Tissue_Type.
A non-negative integer, indicating the maximum number of days that the Collection_Date of a TISSUE_DATA row with this Tissue_Type is allowed to exceed the source individual's BIOGRAPH.Statdate, if any.
This column may be NULL, indicating that the row's
Tissue_Type can be collected any number of days after the
Statdate.
The possible relationships between baboon groups and sleeping groves; whether there is such a relationship and if so whether it the group descended from the grove or ascended into it.
The system uses the ADN column value to enforce data integrity rules.
ADCODES defines values for SWERB_LOC_DATA.ADcode.
Code used to designate the existence and type of relationship between groups and sleeping groves.
This column cannot be changed and must not be
NULL.
The values A
and D are used by
the SWERB_UPLOAD view. These 2 codes must
require SWERB_LOC_DATA.ADtime values -- the
ADCODES.Time values must be
TRUE.[239]
The value N is
used by the SWERB_UPLOAD view when
recording a drinking event.
The values of this column have special meaning to the system. The allowed values are:
AThis ADcode indicates that the group has ascended into a sleeping grove.
DThis ADcode indicates that the group has descended from a sleeping grove.
NThis ADcode indicates that the landscape feature was not used as a sleeping grove.
This column cannot be changed and must not be
NULL.[240]
The values of this column have special meaning to the system. The allowed values are:
TRUEThe related SWERB_LOC_DATA rows must have non-
NULLADtime values.FALSEThe related SWERB_LOC_DATA rows must have
NULLADtime values.
This column may not be NULL.
The different kinds of landscape features.
Note
This table exists to allow for landmarks other than groves and waterholes.
The following special codes can only be altered by suitably privileged individuals. See Special Values.
Special PLACE_TYPES codes
WCode used for a waterhole or rain pool.
GCode used for a grove.
The different kinds of predators that may be seen in the field.
PREDATORS defines values for SWERB_DATA.Predator.
Code identifying the type[241] of predator.
This support table lists the possible confidence scores used when analyzing the accuracy of locations noted in SWERB data.[242]
SWERB_LOC_CONFIDENCES defines values for SWERB_LOC_DATA_CONFIDENCES.Confidence.
Code used to indicate confidence in the accuracy of an observation of a location.
This support table lists the possible different statuses for the observations of specified loacations in SWERB data.
SWERB_LOC_STATUSES defines values for SWERB_LOC_DATA.Loc_Status.
The code C means
certain. It is the default used by the SWERB_UPLOAD when there is no other indication
of certainty.
The code P means
probable. It used by the SWERB_UPLOAD
when there is an indication in the data that the sleeping
grove is not identified with 100% certainty.
The different sources from which times or time estimates were obtained of the SWERB daily group observation starting and ending times. These are the times used when each group began to be observed for the day and when observation of the group was finished for the day when, for some reason, begin or end times were not recorded directly in SWERB.
The code G is
used by the SWERB_UPLOAD view when
uploading data obtained from the GPS units in the
field.
The code
NR indicates that there is no
record of any time source. When this code is used the
related time value must be NULL and the estimated time
flag must be FALSE. See the
explanation in the SWERB_BES
documentation for further detail.
The different sources from which UTM XY coordinates were obtained for landscape features.
SWERB_XYSOURCES defines values for SWERB_GW_LOC_DATA.XYSource.
The value quad is
prohibited from use because this value is used by the SWERB_GW_LOCS view as a XYSource value and intermingled
there with SWERB_XYSources.XYSource values.
The different programs used to retrieve data from the digital weather instrument. Important notes about a program's strengths or weaknesses (e.g. "this program only records its data as integers") should also be noted here.
Note
As discussed earlier , this table has been renamed from its original name: WEATHERHAWK_SOFTWARES.
WEATHER_SOFTWARES defines values for the DIGITAL_WEATHER.WSoftware column.
The different weather stations from which meteorological data are obtained. A weather station can be a collection of instruments at a single location or a single instrument. The content of the table therefore determines whether each WSTATIONS row represents a physical location or a particular instrument. See the content of the table and the Protocol for Data Management: Amboseli Baboon Project for an explanation of the existing practice.[243]
Note
It is a matter of usage whether an existing WSTATIONS code is retired and a new one created when replacing an instrument, or whether the existing code is re-used. See the Protocol for Data Management: Amboseli Baboon Project.
In the XYLoc column, this
table provides the option to record X and Y WGS
1984 UTM Zone 37South
coordinates for the weather station. When such coordinates
are recorded, the source of these coordinates must also be
recorded, in the Loc_Source
column. That is, the XYLoc and
Loc_Source columns must both be
NULL, or both non-NULL.
Tip
To convert an XYLoc value into discrete X and Y coordinates, use the ST_X() and ST_Y() functions, respectively.
To create a new XYLoc value from known X and Y coordinates, use the bb_makepoint() function.
The X and Y WGS 1984 UTM Zone 37South coordinates of this weather station.
This column may be NULL, when no such coordinates are
known or available.
The possible reasons why a behavior gap ended.
GAP_END_STATUSES defines values for the Gap_End_Status column of BEHAVE_GAPS.
[227] Although Babase allows for a hierarchy of body parts it does not require one, and in fact has no support for querying such a hierarchy in a fashion similar to that for the group hierarchy supported by the MEMBERS.Supergroup column. The Bodyregion column can be used as a simple classification column supporting a single level of body part aggregation.
[228] Separate by definition, not necessarily separate in terms of contents. In practice, some people are likely to be listed on both tables.
[229] Silly you. You thought you knew already.
[230] Except by suitably privileged individuals. See Special Values.
[231] As is indeed the case as of this writing.
[232] There is no code for female because at present the protocols do not require one.
[233] ...but can be any number of days after it.
[234] ...and also allows any number of days before it.
[235] For example, perhaps the individual has no MATUREDATES.Matured because they are young and haven't yet matured, or they're a male who dispersed or died before maturity.
[236] baby
[237] “Hardcoded into” to use a term-of-art.
[238] It's not necessary to go all the way back to the SI standard unit. If all your concentrations tend to be in nanomolar with the occasional picomolar, then both units could use nanomolar as their Reference and never bother going back to molar.
[239] Otherwise missing SWERB_LOC_DATA.ADtime values might not be detected in those cases where the data entry protocol calls for 2 waypoints to record sleeping grove information but only 1 waypoint was entered.
[240] In the unlikely event that this column must be changed the following procedure can be followed: Create a new, temporary, ADCODES value with the changed ADN column. Update the SWERB_LOC_DATA table to use the new code. Delete the old code. Re-create the old code with the desired ADN value. Again update SWERB_LOC_DATA to use the re-created code. Delete the temporary ADCODES value.
Because it is unlikely that existing ADCODES.ADN values will need to change it was not thought worthwhile to do the work involved in adding the integrity checking rules to the database which would allow the ADN value to be changed.
[241] These divisions into different "types" could be subdivided separated by species, genus, or something more arbitrary. A decision for data managers.
[242] See SWERB_LOC_DATA_CONFIDENCES for more info about these analyses.
[243] At some future time it may be desirable to extend the database by adding a location code to the WSTATIONS table. This would allow for aggregation of weather station data by location. A number of problems would have to be resolved first, notably what constitutes a location and how to reconcile any differences in the weather station instrumentation. There would also have to be a need. In the meantime simplicity is the best choice.
Table of Contents
- Group Membership and Life Events
- CENSUS_DEMOG (CENSUS extended with DEMOG information)
- CENSUS_DEMOG_SORTED (CENSUS_DEMOG, Sorted)
- CYCPOINTS_CYCLES (CYCPOINTS extended with CYCLES information)
- CYCPOINTS_CYCLES_SORTED (CYCPOINTS_CYCLES, Sorted)
- DEMOG_CENSUS (DEMOG, showing CENSUS information)
- DEMOG_CENSUS_SORTED (DEMOG_CENSUS, Sorted)
- GROUPS_HISTORY
- PARENTS
- POTENTIAL_DADS (Potential Dads)
- PROPORTIONAL_RANKS (RANKS extended with calculated PROPORTIONAL ranks)
- Physical Traits
- ESTROGENS
- GLUCOCORTICOIDS
- HORMONE_PREPS
- HORMONE_RESULTS
- HORMONE_SAMPLES
- HYBRIDMORPH_SCORES
- PROGESTERONES
- TESTOSTERONES
- THYROID_HORMONES
- WOUNDSPATHOLOGIES (All Wound/Pathology Data, Together)
- WP_DETAILS_AFFECTEDPARTS (WP_DETAILS, extended with WP_AFFECTEDPARTS)
- WP_HEALS (WP_HEALUPDATES, extended)
- WP_REPORTS_OBSERVERS (WP_REPORTS, extended with WP_OBSERVERS)
- Sexual Cycles
- CYCLES_SEXSKINS (CYCLES extended with SEXSKINS information)
- CYCLES_SEXSKINS_SORTED (CYCLES_SEXSKINS, Sorted)
- MATERNITIES (completed reproductive events)
- MTD_CYCLES (CYCLES and Mdate, Tdate, and Ddate CYCPOINTS data)
- SEXSKINS_CYCLES (CYCLES extended with SEXSKINS information)
- SEXSKINS_CYCLES_SORTED (SEXSKINS_CYCLES, Sorted)
- SEXSKINS_REPRO_NOTES (SEXSKINS extended with REPRO_NOTES)
- Social and Multiparty Interactions
- ACTOR_ACTEES (Complete social interactions, INTERACT_DATA extended twice with PARTS)
- INTERACT (INTERACT_DATA, with enhanced dates and times)
- INTERACT_SORTED
- MPI_EVENTS (Dyadic social interactions that comprise multiparty interaction collections, MPIS joined with MPI_DATA extended twice with MPI_PARTS)
- MPI_UPLOAD: Upload Multiparty Interactions
- POINTS (POINT_DATA, with enhanced times)
- POINTS_SORTED (POINTS, Sorted)
- SAMPLES_GOFF (SAMPLES, with the Group OF the Focal)
- Darting
- ANESTH_STATS (darting additional Anesthetic Statistics)
- BODYTEMP_STATS (darting Body Temperature Statistics)
- CHEST_STATS (darting Chest circumference Statistics)
- CROWNRUMP_STATS (darting Crown-to-Rump Statistics)
- DART_FLOW_CYTOMETRY_UPLOAD (facility for recording flow cytometry data)
- DART_LOGISTICS_UPLOAD (facility for adding data about the logistics of a darting)
- DART_MORPHOLOGY_UPLOAD (facility for adding morphology data collected during a darting)
- DART_PHYSIOLOGY_UPLOAD (facility for adding physiology data collected during a darting)
- DART_SAMPLES_UPLOAD (facility for recording tissue samples collected during a darting)
- DART_TEETH_UPLOAD (facility for adding tooth data collected during a darting)
- DART_TESTES_ARC_UPLOAD (facility for adding testicle circumference data)
- DART_TESTES_DIAM_UPLOAD (facility for adding testicle diameter data)
- DART_TICKS_UPLOAD (facility for adding data from parasite counts performed during a darting)
- DART_VAGINAL_PHS_UPLOAD (facility for recording vaginal pH measurements performed during a darting)
- DART_WBC_COUNTS_UPLOAD (facility for recording white blood cell counts from blood smears)
- DSAMPLES (darting sample records with columns for each sample type)
- DENT_CODES (darting Dentition records with columns for each Toothcode)
- DENT_SITES (darting Dentition records with columns for each Toothsite)
- HUMERUS_STATS (darting Humerus length Statistics)
- PCV_STATS (darting PCV Statistics)
- TESTES_ARC_STATS (darting Testes circumference Statistics)
- TESTES_DIAM_STATS (darting Testes Diameter Statistics)
- ULNA_STATS (darting Ulna length Statistics)
- VAGINAL_PH_STATS (darting Vaginal pH Statistics)
- Inventory
- SWERB Data (Group-level Geolocation Data)
- QUADS (map Quadrants)
- SWERB (Group level gps point samples)
- SWERB_DATA_XY (The SWERB_DATA table with separate X and Y coordinates)
- SWERB_DEPARTS (SWERB observation team Departures from camp)
- SWERB_GW_LOCS (SWERB Grove and Waterhole Locations)
- SWERB_GW_LOC_DATA_XY (The SWERB_GW_LOC_DATA table with separate X and Y coordinates)
- SWERB_LOC_GPS_XY (The SWERB_LOC_GPS table with separate X and Y coordinates)
- SWERB_LOCS (placement of a group at a landscape feature)
- SWERB_UPLOAD (facility for uploading data into SWERB)
- Weather Data
- Views Which Add Gid To Tables
- The BIRTH_GRP View
- The ENTRYDATE_GRP View
- The STATDATE_GRP View
- The CONSORTDATES_GRP View
- The CYCGAPDAYS_GRP View
- The CYCGAPS_GRP View
- The CYCSTATS_GRP View
- The DARTINGS_GRP View
- The DISPERSEDATES_GRP View
- The MATUREDATES_GRP View
- The MDINTERVALS_GRP View
- The MMINTERVALS_GRP View
- The RANKDATES_GRP View
- The REPSTATS_GRP View
The documentation of each view contains a short description of the purpose of the view, the query used to generate the view, a diagram of the Babase tables contained in the view, a table showing the columns contained in the view, and notes on the operations (INSERT, UPDATE, or DELETE) allowed on the view. For further information on the columns' content see the documentation of each column in the table that is the source of the view's data.
Note
Babase contains schemas that use views to
organize the Babase content. The views in these schemas refer
to tables or views within the babase schema and
are not otherwise documented.
Warning
Attempts to update “computed columns”, columns that appear in the view but not in the underlying data tables, may be silently ignored. This is also sometimes true of actual data columns that are expected to automatically have their values assigned by Babase. Changes that are silently ignored produce no error message. The ignored changes are not made at the same time that changes to other columns are made.
The views are being changed as time permits so that there are no cases where errors are silently ignored.
The documentation of each view describes which columns can not be changed through the view.
The entity-relationship diagrams which document each view use the same key as this documents other entity-relationships diagrams. The key is show in Figure 2.1: “Key to the Babase Entity Relationship Diagrams”.
Note
If you have trouble viewing the diagrams in your browser, you may wish to view them in PDF format. The diagrams are available in The Babase Pocket Reference (approx. 4.8MB) in PDF form.
There are two differences between the entity-relationship diagrams which document the views and those which show the relationship between the Babase tables. First, the ER diagrams of the tables are a complete reference, they show all of each table's columns. The ER diagrams of the views show only those columns used in the view. Second, the view ER diagrams follow the column names of each Babase table with parenthesis that contain the name each column takes in the view.[244]
Contains one row for every row in CENSUS. Each row contains the CENSUS columns and the related DEMOG columns. In those cases where there is a
CENSUS row but no related DEMOG row the DEMOG columns will
be NULL. Because there is a one-to-one relationship between
CENSUS and DEMOG, and a
DEMOG row always has a related CENSUS row, there is little utility in maintaining
the DEMOG row without maintaining the
related CENSUS row. This view provides a
convenient way to maintain the CENSUS/DEMOG combination.
Figure 6.1. Query Defining the CENSUS_DEMOG View
SELECT census.cenid AS cenid
, census.sname AS sname
, census.date AS date
, census.grp AS grp
, census.status AS status
, census.cen AS cen
, demog.reference AS reference
, demog.comment AS comment
FROM census LEFT OUTER JOIN demog ON (census.cenid = demog.cenid);
Table 6.1. Columns in the CENSUS_DEMOG View
| Column | From | Description |
|---|---|---|
| Cenid | CENSUS.Cenid | Unique identifier of the CENSUS row. |
| Sname | CENSUS.Sname | Individual who's location has been recorded. |
| Date | CENSUS.Date | Date of demography note. |
| Grp | CENSUS.Grp | Group where the individual was located. |
| Status | CENSUS.Status | Source of location information. When the source is both a demography note and another source, like a census, the other source is shown. |
| Cen | CENSUS.Cen | Whether or not there was an entry on the field census data sheet for the individual on the given date. |
| Reference | DEMOG.Reference | The group identifying the written field notebook where the demography note can be found. |
| Comment | DEMOG.Comment | The demography note text. |
Inserting a row into CENSUS_DEMOG inserts two rows,
one into CENSUS and one into DEMOG, as expected. However, if the
underlying DEMOG columns are NULL, no
DEMOG row will be inserted.
Warning
The PostgreSQL nextval()
function cannot be part of an INSERT
expression which assigns a value to this view's Cenid
column.
Updating a row in CENSUS_DEMOG updates the underlying columns in CENSUS and DEMOG, as expected. However, the relationship between CENSUS and DEMOG introduces some complications.
Caution
The CENSUS table is updated before the DEMOG table. Because updating the DEMOG table can change the CENSUS.Status column the resulting value may not be that specified by the update.
Updating the Cenid column updates[245] the Cenid columns in both CENSUS and DEMOG. Setting
all the DEMOG columns (Cenid excepted) to NULL causes the
deletion of the DEMOG row. Setting
DEMOG columns to a non-NULL value
when all the DEMOG columns were NULL
previously creates a new row in DEMOG.
Caution
The CENSUS-DEMOG view cannot be used to delete arbitrary CENSUS rows.
Deleting rows from CENSUS_DEMOG updates the database in a fashion that removes the related demography note information from storage.
Deleting a row in CENSUS_DEMOG deletes the
underlying row in CENSUS when
appropriate; when the CENSUS row exists
only because there is an underlying row in DEMOG. That is, the CENSUS
row is deleted if and only if the CENSUS.Cen column is
FALSE. If there is an underlying row in DEMOG it is always deleted.
Contains one row for every row in the CENSUS_DEMOG view. The only difference between this view and the CENSUS_DEMOG view is that this view is sorted.
Figure 6.3. Query Defining the CENSUS_DEMOG_SORTED View
SELECT census.cenid AS cenid
, census.sname AS sname
, census.date AS date
, census.grp AS grp
, census.status AS status
, census.cen AS cen
, demog.reference AS reference
, demog.comment AS comment
FROM census LEFT OUTER JOIN demog ON (census.cenid = demog.cenid)
ORDER BY census.sname, census.date;
Table 6.2. Columns in the CENSUS_DEMOG_SORTED View
| Column | From | Description |
|---|---|---|
| Cenid | CENSUS.Cenid | Unique identifier of the CENSUS row. |
| Sname | CENSUS.Sname | Individual who's location has been recorded. |
| Date | CENSUS.Date | Date of demography note. |
| Grp | CENSUS.Grp | Group where the individual was located. |
| Status | CENSUS.Status | Source of location information. When the source is both a demography note and another source, like a census, the other source is shown. |
| Cen | CENSUS.Cen | Whether or not there was an entry on the field census data sheet for the individual on the given date. |
| Reference | DEMOG.Reference | The group identifying the written field notebook where the demography note can be found. |
| Comment | DEMOG.Comment | The demography note text. |
The operations allowed are as described in the CENSUS_DEMOG view.
Contains one row for every row in CYCPOINTS. Each row contains the CYCPOINTS columns and the related CYCLES columns. Because there is a many-to-one relationship between CYCPOINTS and CYCLES, the same CYCLES data will appear repeatedly, once for each related CYCPOINTS row. As a CYCPOINTS row always has a related CYCLES row, and the CYCLES row is what identifies the cycling female, when working with the CYCPOINTS table alone it is difficult to tell which dates belong to which females. This view provides a convenient way to create and maintain the CYCPOINTS/CYCLES combination.
Figure 6.5. Query Defining the CYCPOINTS_CYCLES View
SELECT cycles.cid AS cid
, cycles.sname AS sname
, cycles.seq AS seq
, cycles.series AS series
, cycpoints.cpid AS cpid
, cycpoints.date AS date
, cycpoints.edate AS edate
, cycpoints.ldate AS ldate
, cycpoints.code AS code
, cycpoints.source AS source
FROM cycles, cycpoints
WHERE cycles.cid = cycpoints.cid;
Table 6.3. Columns in the CYCPOINTS_CYCLES View
| Column | From | Description |
|---|---|---|
| Cid | CYCLES.Cid | Arbitrary number uniquely identifying the CYCLES row. |
| Sname | CYCLES.Sname | Female that is cycling. |
| Seq | CYCLES.Seq (readonly) | Number indicating the cycle's position in time within the sequence of all the cycles of the female. Counts from 1 upwards. |
| Series | CYCLES.Series (readonly) | Number indicating with which CYCLES (Female Sexual Cycles) of continuous observation the cycle belongs. |
| Cpid | CYCPOINTS.Cpid | Number uniquely identifying the CYCPOINTS row. |
| Date | CYCPOINTS.Date | Date-of-record of the sexual cycle transition event. |
| Edate | CYCPOINTS.Edate | Earliest possible date for the sexual cycle transition event. |
| Ldate | CYCPOINTS.Ldate | Latest possible date for the sexual cycle transition event. |
| Code | CYCPOINTS.Code | The type of sexual cycle transition event. Mdate, Tdate, or Ddate. |
| Source | CYCPOINTS.Source | Code indicating from whence the data was derived. This has a bearing as to its accuracy. |
Tip
In most cases Cid, Cpid, Seq, and Series should be
unspecified (or specified as NULL), in which case Babase
will compute and assign the correct values.
Inserting a row into CYCPOINTS_CYCLES inserts a row into CYCPOINTS, as expected. Whether or not a row is inserted into CYCLES depends on whether or not the new CYCPOINTS row should be associated with a new CYCLES row or an existing one. When a Cid is supplied and a CYCLES row already exists with the given Cid then the underlying CYCLES row is updated to conform with the inserted data. When a Cid is supplied and Babase finds that the underlying CYCPOINTS row should be related to a CYCLES row with a different Cid the system silently ignores the supplied Cid.
Contains one row for every row in the CYCPOINTS_CYCLES view. This view is sorted for ease of maintenance.
Figure 6.7. Query Defining the CYCPOINTS_CYCLES_SORTED View
SELECT cycles.cid AS cid
, cycles.sname AS sname
, cycles.seq AS seq
, cycles.series AS series
, cycpoints.cpid AS cpid
, cycpoints.date AS date
, cycpoints.edate AS edate
, cycpoints.ldate AS ldate
, cycpoints.code AS code
, cycpoints.source AS source
FROM cycles, cycpoints
WHERE cycles.cid = cycpoints.cid
ORDER BY cycles.sname, cycpoints.date;
Table 6.4. Columns in the CYCPOINTS_CYCLES_SORTED View
| Column | From | Description |
|---|---|---|
| Cid | CYCLES.Cid | Arbitrary number uniquely identifying the CYCLES row. |
| Sname | CYCLES.Sname | Female that is cycling. |
| Seq | CYCLES.Seq (readonly) | Number indicating the cycle's position in time within the sequence of all the cycles of the female. Counts from 1 upwards. |
| Series | CYCLES.Series (readonly) | Number indicating with which CYCLES (Female Sexual Cycles) of continuous observation the cycle belongs. |
| Cpid | CYCPOINTS.Cpid | Number uniquely identifying the CYCPOINTS row. |
| Date | CYCPOINTS.Date | Date-of-record of the sexual cycle transition event. |
| Edate | CYCPOINTS.Edate | Earliest possible date for the sexual cycle transition event. |
| Ldate | CYCPOINTS.Ldate | Latest possible date for the sexual cycle transition event. |
| Code | CYCPOINTS.Code | The type of sexual cycle transition event. Mdate, Tdate, or Ddate. |
| Source | CYCPOINTS.Source | Code indicating from whence the data was derived. This has a bearing as to its accuracy. |
The operations allowed are as described in the CYCPOINTS_CYCLES view.
Contains one row for every row in DEMOG. Each row contains the DEMOG columns and the related CENSUS columns. In those cases where there is a CENSUS row but no related DEMOG row no row exists. Because there is a one-to-one relationship between CENSUS and DEMOG and a DEMOG row always has a related CENSUS row, and because without the CENSUS information it is difficult to tell to which individual the demography note refers , there is little utility in maintaining the DEMOG row without maintaining the related CENSUS row. This view provides a convenient way to maintain the CENSUS/DEMOG combination.
Note
The DEMOG_CENSUS view is very similar to the CENSUS_DEMOG view. It is unclear which is more useful so both exist.
Figure 6.9. Query Defining the DEMOG_CENSUS View
SELECT census.cenid AS cenid
, census.sname AS sname
, census.date AS date
, census.grp AS grp
, census.status AS status
, census.cen AS cen
, demog.reference AS reference
, demog.comment AS comment
FROM census, demog
WHERE census.cenid = demog.cenid;
Table 6.5. Columns in the DEMOG_CENSUS View
| Column | From | Description |
|---|---|---|
| Cenid | CENSUS.Cenid | Unique identifier of the CENSUS row. |
| Sname | CENSUS.Sname | Individual who's location has been recorded. |
| Date | CENSUS.Date | Date of demography note. |
| Grp | CENSUS.Grp | Group where the individual was located. |
| Status | CENSUS.Status | Source of location information. When the source is both a demography note and another source, like a census, the other source is shown. |
| Cen | CENSUS.Cen | Whether or not there was an entry on the field census data sheet for the individual on the given date. |
| Reference | DEMOG.Reference | The group identifying the written field notebook where the demography note can be found. |
| Comment | DEMOG.Comment | The demography note text. |
Inserting a row into DEMOG_CENSUS inserts a row,
into DEMOG, as expected, but only when
the underlying DEMOG columns are not
NULL. If the values of the columns belonging to the
DEMOG table are null then the insert
raises an error.
A new CENSUS row is inserted if
there is not already an existing CENSUS
row, otherwise the existing CENSUS row
is updated. To leave the value of an existing CENSUS row untouched either omit the column
from the insert or specify the NULL value.
Warning
Values must be supplied for either the Cenid or all of the Sname, Date, and Grp columns or else the system will be unable to identify pre-existing CENSUS rows updated by inserts to DEMOG_CENSUS. The system may silently ignore insert operations when too few column values are supplied.
When a new CENSUS row is created the Cen column defaults to false.
Warning
The PostgreSQL nextval()
function cannot be part of an INSERT
expression which assigns a value to this view's Cenid
column.
Updating a row in DEMOG_CENSUS updates the underlying columns in CENSUS and DEMOG, as expected. However, the relationship between CENSUS and DEMOG introduces some complications.
Caution
The CENSUS table is updated before the DEMOG table. Because updating the DEMOG table can change the CENSUS.Status column the resulting value may not be that specified by the update.
Updating the Cenid column updates[246] the Cenid columns in both CENSUS and DEMOG.
Deleting rows from DEMOG_CENSUS updates the database in a fashion that removes the related demography note information from storage.[247]
Deleting a row in DEMOG_CENSUS deletes the
underlying row in CENSUS when
appropriate; when the CENSUS row exists
only because there is an underlying row in DEMOG. That is, the CENSUS
row is deleted if and only if the CENSUS.Cen column is
FALSE. If there is an underlying row in DEMOG it is always deleted.
Note
Regardless of whether the underlying CENSUS row is deleted a delete operation will always cause the deleted row to disappear from the DEMOG_CENSUS view.
Contains one row for every row in the DEMOG_CENSUS view. The only difference between this view and the DEMOG_CENSUS view is that this view is sorted.
Figure 6.11. Query Defining the DEMOG_CENSUS_SORTED View
SELECT census.cenid AS cenid
, census.sname AS sname
, census.date AS date
, census.grp AS grp
, census.status AS status
, census.cen AS cen
, demog.reference AS reference
, demog.comment AS comment
FROM census, demog
WHERE census.cenid = demog.cenid
ORDER BY census.sname, census.date;
Table 6.6. Columns in the DEMOG_CENSUS_SORTED View
| Column | From | Description |
|---|---|---|
| Cenid | CENSUS.Cenid | Unique identifier of the CENSUS row. |
| Sname | CENSUS.Sname | Individual who's location has been recorded. |
| Date | CENSUS.Date | Date of demography note. |
| Grp | CENSUS.Grp | Group where the individual was located. |
| Status | CENSUS.Status | Source of location information. When the source is both a demography note and another source, like a census, the other source is shown. |
| Cen | CENSUS.Cen | Whether or not there was an entry on the field census data sheet for the individual on the given date. |
| Reference | DEMOG.Reference | The group identifying the written field notebook where the demography note can be found. |
| Comment | DEMOG.Comment | The demography note text. |
The operations allowed are as described in the DEMOG_CENSUS view.
Contains one row for every row in the GROUPS table. This view portrays a group's history in a more-accessible format than the GROUPS table. It collects into one row all the dates that are relevant to a particular group, including (in the case of groups that fissioned) the date they became "impermanent" by starting to fission.
This view is similar to GROUPS, but omits columns used predominantly for data entry and validation. This view also renames some columns for clarity, and adds three calculated columns.
Figure 6.13. Query Defining the GROUPS_HISTORY View
SELECT groups.gid AS gid
, groups.name AS name
, groups.from_group AS from_group
, groups.to_group AS to_group
, CASE
WHEN groups.from_group IS NULL
AND NOT EXISTS (SELECT 1
FROM groups AS from_groups
WHERE from_groups.to_group = groups.gid)
THEN groups.permanent
ELSE groups.start
END AS first_observed
, CASE
WHEN groups.study_grp IS NULL
THEN NULL
WHEN groups.from_group IS NULL
AND NOT EXISTS (SELECT 1
FROM groups AS from_groups
WHERE from_groups.to_group = groups.gid)
THEN groups.permanent
ELSE (SELECT date
FROM census
WHERE census.grp = groups.gid
AND census.cen
ORDER BY date
LIMIT 1)
END AS first_study_grp_census
, groups.permanent AS permanent
, (SELECT descgroups_start.start
FROM babase.groups AS descgroups_start
WHERE descgroups_start.from_group = groups.gid
OR descgroups_start.gid = groups.to_group
ORDER BY descgroups_start.start
LIMIT 1
) AS impermanent
, groups.cease_to_exist AS cease_to_exist
, groups.last_reg_census AS last_reg_census
, groups.study_grp
FROM babase.groups;
Table 6.7. Columns in the GROUPS_HISTORY View
| Column | From | Description |
|---|---|---|
| Gid | GROUPS.Gid | Unique identifier of the GROUPS row. |
| Name | GROUPS.Name | Name of this group. |
| From_Group | GROUPS.From_group | The Gid of the group from which this group was created. |
| First_Observed | GROUPS.Permanent, GROUPS.Start | The first date the group was observed. For groups that are fission or fusion products of other known groups, this is the group's Start. Otherwise, this is the group's Permanent. |
| First_Study_Grp_Census | GROUPS.Permanent, CENSUS.Date | The first date that a study group was
observed as its own group and not a subgroup from
its parent group (in the case of fissions) or as a
temporary multi-group encounter (in the case of
fusions). For non-study groups (Study_Grp is NULL), this is
NULL. For groups of unknown lineage (groups whose
From_group is NULL and
whose Gid does not exist as
a To_group)[a], this is the group's Permanent date. Otherwise, this
is the group's earliest CENSUS.Date
where Cen is
TRUE. |
| Permanent | GROUPS.Permanent | The first date on which the group was recognized as its own distinct group. |
| Impermanent | GROUPS.Start | The earliest Start date of this group's fission or fusion products. |
| Cease_To_Exist | GROUPS.Cease_To_Exist | The last date of this group's existence, and the day before fission or fusion products of this group became permanent. |
| Last_Reg_Census | GROUPS.Last_Reg_Census | The date of the last regular census done on the group. |
| Study_Grp | GROUPS.Study_Grp | The date the group became an "official" study
group, or NULL if the group was never a study
group. |
[a] Which is expected to only occur when a previously unseen group is first seen and becomes a known group. | ||
Contains one row for every BIOGRAPH
row for which there is either a row in MATERNITIES with a record of the individual's
mother or there is a row in DADS_CONSENSUS
with a record of the individual's father -- where DADS_CONSENSUS.Dad_Consensus is not NULL.
Note
A row in this view can have a NULL Mom or a NULL
Dad, but not both. When there is neither a Mom (i.e., the
offspring has a NULL BIOGRAPH.Pid) nor a Dad (i.e., the offspring has
either a NULL.DADS_CONSENSUS.Dad_Consensus or no related DADS_CONSENSUS data row for the father at all)
the view has no row for the individual.
Figure 6.15. Query Defining the PARENTS View
SELECT biograph.sname AS kid
, maternities.mom AS mom
, dads_consensus.dad_consensus AS dad
, maternities.zdate AS zdate
, maternities.zdate_grp AS momgrp
, members.grp AS dadgrp
FROM biograph
LEFT OUTER JOIN maternities
ON (maternities.child = biograph.sname)
LEFT OUTER JOIN dads_consensus
ON (dads_consensus.kid = biograph.sname)
LEFT OUTER JOIN members
ON (members.sname = dads_consensus.dad_consensus
AND members.date = maternities.zdate)
WHERE maternities.mom IS NOT NULL
OR dads_consensus.dad_consensus IS NOT NULL;
Table 6.8. Columns in the PARENTS View
| Column | From | Description |
|---|---|---|
| Kid | BIOGRAPH.Sname | Identifier (Sname) of the offspring. |
| Mom | CYCLES.Sname | Identifier (Sname)
of the mother, or NULL if the mother is not
known. |
| Dad | DADS_CONSENSUS.Dad_Consensus | Identifier (Sname)
of the father -- the manually chosen
father-of-choice --, or NULL if there is
none. |
| Zdate | CYCPOINTS.Date | Conception date-of-record, or NULL if the
mother is not known. |
| Momgrp | MEMBERS.Grp | Mother's group as of the conception
date-of-record, or NULL if the mother is not
known. |
| Dadgrp | MEMBERS.Grp | The group of the father on the Zdate, or
NULL if there is either no consensus dad or the
Zdate is not known. |
Contains one row for every (completed) female reproductive event for every male more than 2192 days old (approximately 6 years) present in the mother's supergroup during her fertile period. So, one row for every potential dad of every birth and fetal loss. The Potential_Dads-Status column can be used to distinguish males that are adult on the Zdate from subadults from males that have no record of testicular enlargement -- the males having no MATUREDATES.Matured.
Figure 6.17. Query Defining the POTENTIAL_DADS View
SELECT maternities.child_bioid AS bioid
, maternities.child AS kid
, maternities.mom AS mom
, maternities.zdate AS zdate
, maternities.zdate_grp AS grp
, pdads.sname AS pdad
, CASE
WHEN rankdates.ranked <= maternities.zdate
THEN 'A'
WHEN maturedates.matured <= maternities.zdate
THEN 'S'
ELSE 'O'
END
AS status
, maternities.zdate - pdads.birth AS pdad_age_days
, trunc((maternities.zdate - pdads.birth) / 365.25, 1)
AS pdad_age_years
, (SELECT count(*)
FROM members as dadmembers
JOIN members AS mommembers
ON (mommembers.date = dadmembers.date
AND mommembers.supergroup = dadmembers.supergroup)
WHERE dadmembers.sname = pdads.sname
AND dadmembers.date < maternities.zdate
AND dadmembers.date >= maternities.zdate - 5
AND mommembers.sname = maternities.mom
AND mommembers.date < maternities.zdate
AND mommembers.date >= maternities.zdate - 5)
AS estrous_presence
, (SELECT count(*)
FROM actor_actees
WHERE actor_actees.date < maternities.zdate
AND actor_actees.date >= maternities.zdate - 5
AND (actor_actees.act = 'M'
OR actor_actees.act = 'E')
AND actor_actees.actor = pdads.sname
AND actor_actees.actee = maternities.mom)
AS estrous_me
, (SELECT count(*)
FROM actor_actees
WHERE actor_actees.date < maternities.zdate
AND actor_actees.date >= maternities.zdate - 5
AND actor_actees.act = 'C'
AND actor_actees.actor = pdads.sname
AND actor_actees.actee = maternities.mom)
AS estrous_c
FROM maternities
JOIN biograph AS pdads
ON (pdads.sname
IN (SELECT dadmembers.sname
FROM members AS dadmembers
JOIN members AS mommembers
ON (mommembers.date = dadmembers.date
AND mommembers.supergroup
= dadmembers.supergroup)
WHERE dadmembers.sname = pdads.sname
AND dadmembers.date < maternities.zdate
AND dadmembers.date >= maternities.zdate - 5
AND mommembers.sname = maternities.mom
AND mommembers.date < maternities.zdate
AND mommembers.date >= maternities.zdate - 5))
LEFT OUTER JOIN rankdates
ON (rankdates.sname = pdads.sname)
LEFT OUTER JOIN maturedates
ON (maturedates.sname = pdads.sname)
WHERE pdads.sex = 'M'
-- Speed things up by eliminating potential dads
-- who could not possibly interpolate into the mom's group
-- during the fertile period.
AND pdads.statdate >= maternities.zdate - 5 - 14
-- Potential dad must be at least 2192 days old
-- (approximately 6 years) on the zdate.
AND maternities.zdate - pdads.birth >= 2192;
Figure 6.19. Entity Relationship Diagram of that portion of the POTENTIAL_DADS View which places the mother and potential father in the same group during the fertile period
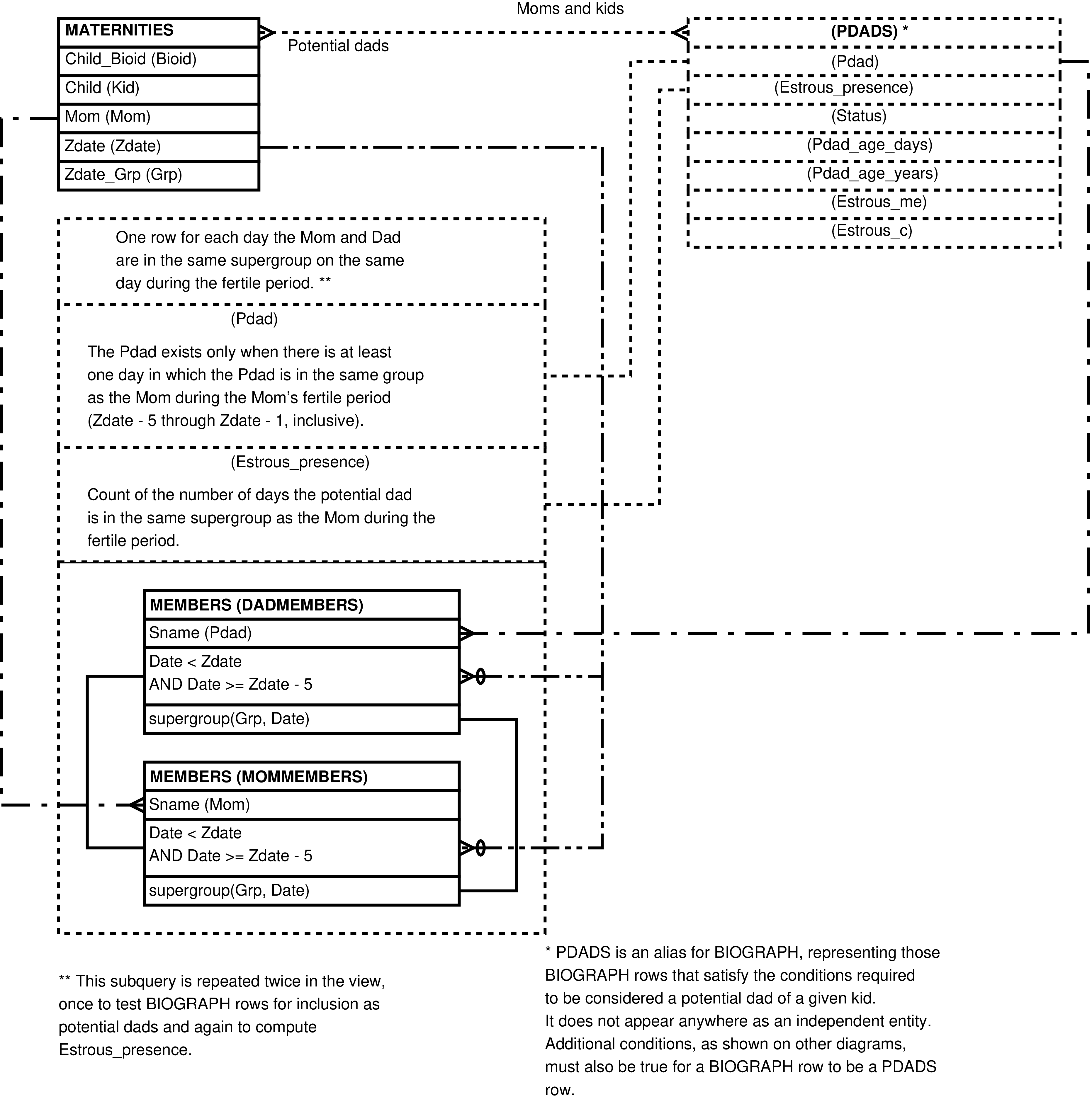 |
Figure 6.20. Entity Relationship Diagram of that portion of the POTENTIAL_DADS View having easily computed columns
 |
Figure 6.21. Entity Relationship Diagram of that portion of the POTENTIAL_DADS View involving social interactions
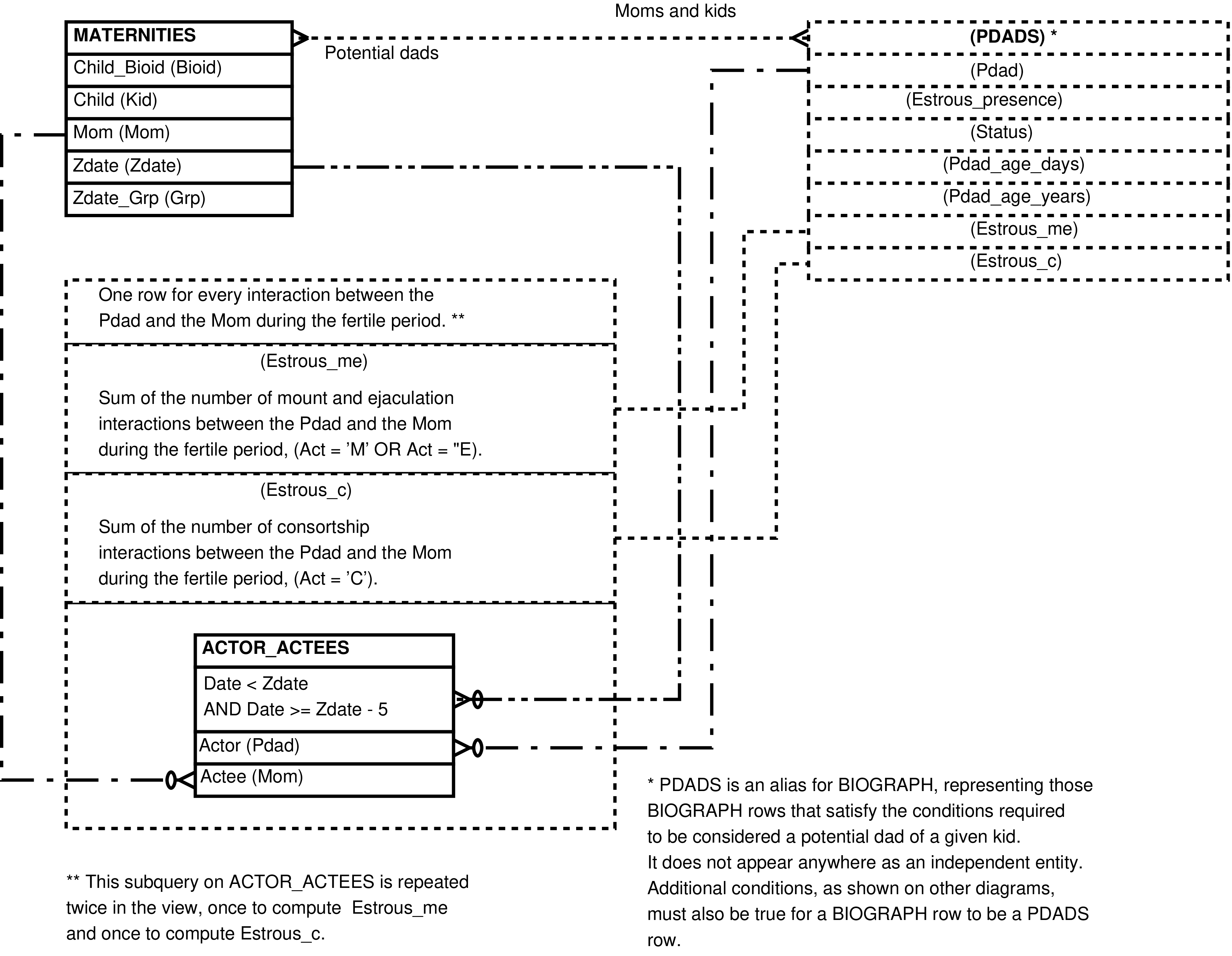 |
Table 6.9. Columns in the POTENTIAL_DADS View
| Column | From | Description |
|---|---|---|
| Bioid | BIOGRAPH.Bioid | Numeric identifier (Bioid) of the offspring. |
| Kid | BIOGRAPH.Sname | Identifier (Sname) of the offspring. |
| Mom | CYCLES.Sname | Identifier (Sname) of the mother. |
| Zdate | CYCPOINTS.Date | Conception date-of-record. |
| Zdate_Grp | MEMBERS.Grp | Mother's group as of the conception date-of-record. |
| Pdad | BIOGRAPH.Sname | Identifier (Sname) of the potential father. |
| Status |
| The maturity of the potential dad as of the
Zdate, as follows:
POTENTIAL_DADS.Status values
|
| Pdad_age_days | Zdate - BIOGRAPH.Birth | The age, in days, of the potential dad as of the Zdate -- the Zdate minus the potential dad's BIOGRAPH.Birth. |
| Pdad_age_years | Pdad_age_days / 365.25 | The age, in years, of the potential dad as of the Zdate. |
| Estrous_presence | Subquery on MEMBERS -- see Figure 6.19. | Count of the number of days the potential dad is in the same supergroup as the supergroup of the mother during the mother's fertile period -- the 5 days prior to the Zdate. |
| Estrous_me | Subquery on ACTOR_ACTEES -- see Figure 6.21. | Sum of the number of mounts and ejaculation interactions between the Mom and the Pdad during the fertile period -- the 5 days prior to the Zdate. |
| Estrous_c | Subquery on ACTOR_ACTEES -- see Figure 6.21. | Sum of the number of consortship interactions between the Mom and the Pdad during the fertile period -- the 5 days prior to the Zdate. |
Contains one row for every row in RANKS. Each row contains all the columns from RANKS and an additional column with the calculated proportional rank.
Proportional rank is a method of ranking that accounts
for the size of the group[248]. Its values should extend between
0 (low rank) and 1 (high
rank), and can be interpreted as "the percent of the group over
which this individual is dominant".
Caution
Be careful when comparing ordinal and proportional rank
values to each other. Ordinal ranks (from RANKS) begin at 1 (high rank),
with ascending values indicating lower rank. Proportional
ranks go in the reverse direction: as proportional rank
values ascend, these values indicate higher ranks.
Figure 6.22. Query Defining the PROPORTIONAL_RANKS View
WITH num_indivs AS (
SELECT ranks.rnkdate
, ranks.grp
, ranks.rnktype
, count(*) AS num_members
FROM ranks
GROUP BY ranks.rnkdate, ranks.grp, ranks.rnktype)
SELECT ranks.rnkid AS rnkid
, ranks.sname AS sname
, ranks.rnkdate AS rnkdate
, ranks.grp AS grp
, ranks.rnktype AS rnktype
, ranks.rank AS ordrank
, ranks.ags_density AS ags_density
, ranks.ags_reversals AS ags_reversals
, ranks.ags_expected AS ags_expected
, CASE
WHEN num_indivs.num_members = 1 THEN 1::numeric
ELSE 1 - ((ranks.rank - 1)::numeric / (num_indivs.num_members - 1)::numeric)
END::numeric(5,4) AS proprank
FROM ranks
JOIN num_indivs
ON (num_indivs.rnkdate = ranks.rnkdate
AND num_indivs.grp = ranks.grp
AND num_indivs.rnktype = ranks.rnktype);
Table 6.10. Columns in the PROPORTIONAL_RANKS View
| Column | From | Description |
|---|---|---|
| RnkId | RANKS.Rnkid | Unique identifier of the RANKS row. |
| Sname | RANKS.Sname | Sname of the ranked individual. |
| Rnkdate | RANKS.Rnkdate | The date indicating the year and month of this ranking. |
| Grp | RANKS.Grp | The group in which this individual is ranked. |
| Rnktype | RANKS.Rnktype | The kind of rank assigned to this individual. |
| OrdRank | RANKS.Rank | The ordinal rank assigned to this individual. |
| PropRank |
| The calculated proportional rank for this individual. Expressed as a value between 0 (low rank) and 1 (high rank), inclusive. |
Contains one row for every sample whose estrogen
concentration has been measured by any kit with a known
correction factor. Results from kits whose HORMONE_KITS.Correction is NULL are
omitted.
Tip
Use this view to see estrogen concentration in all the hormone samples. It joins all the pertinent tables together to gather information, and omits results that are not considered reliable.
This view includes a "Hormone" column that indicates
which hormone was measured in each assay. By definition, this
column will be E in every row,
so it may seem odd to include the row at all. The column is
retained as a courtesy to users, especially for those who
might unify the rows from this view with rows of other,
similar views (e.g. GLUCOCORTICOIDS, PROGESTERONES, etc.).
Figure 6.24. Query Defining the ESTROGENS View
SELECT hormone_sample_data.tid
, hormone_prep_series.hpsid
, hormone_result_data.hrid
, hormone_sample_data.hsid
, biograph.sname
, tissue_data.collection_date
, tissue_data.collection_date_status AS collection_date_status
, hormone_sample_data.fzdried_date AS fzdried_date
, hormone_sample_data.sifted_date AS sifted_date
, meoh_ext.procedure_date AS me_extracted
, spe.procedure_date AS sp_extracted
, hormone_result_data.raw_ng_g AS raw_ng_g
, corrected_hormone(hormone_result_data.raw_ng_g, hormone_kits.correction) AS corrected_ng_g
, hormone_result_data.assay_date
, hormone_kits.hormone AS hormone
, hormone_result_data.kit AS kit
, hormone_sample_data.comments AS sample_comments
, hormone_result_data.comments AS result_comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ
JOIN hormone_prep_series
ON hormone_prep_series.tid = hormone_sample_data.tid
JOIN hormone_result_data
ON hormone_result_data.hpsid = hormone_prep_series.hpsid
JOIN hormone_kits
ON hormone_kits.kit = hormone_result_data.kit
AND hormone_kits.correction IS NOT NULL
AND hormone_kits.hormone = 'E'
LEFT JOIN hormone_prep_data AS meoh_ext
ON meoh_ext.procedure = 'MEOH_EXT'
AND meoh_ext.hpsid = hormone_prep_series.hpsid
LEFT JOIN hormone_prep_data AS spe
ON spe.procedure = 'SPE'
AND spe.hpsid = hormone_prep_series.hpsid;
Table 6.11. Columns in the ESTROGENS View
| Column | From | Description |
|---|---|---|
| TId | HORMONE_SAMPLE_DATA.TId | Identifier of the HORMONE_SAMPLE_DATA row and tissue sample |
| HPSId | HORMONE_PREP_SERIES.HPSId | Identifier of the prep series |
| HRId | HORMONE_RESULT_DATA.HRId | Identifier of the assay that generated this result |
| HSId | HORMONE_SAMPLE_DATA.HSId | User-generated identifier for the tissue sample |
| Sname | BIOGRAPH.Sname | Sname of the individual from whom this sample came |
| Collection_Date | TISSUE_DATA.Collection_Date | Date the tissue sample was collected |
| Collection_Date_Status | TISSUE_DATA.Collection_Date_Status | The status of this Collection_Date |
| FzDried_Date | HORMONE_SAMPLE_DATA.FzDried_Date | Date the sample was freeze-dried |
| Sifted_Date | HORMONE_SAMPLE_DATA.Sifted_Date | Date the freeze-dried sample was sifted |
| ME_Extracted | HORMONE_PREP_DATA.Procedure_Date | Date of methanol extraction, in prep for this result |
| SP_Extracted | HORMONE_PREP_DATA.Procedure_Date | Date of solid-phase extraction, in prep for this result |
| Raw_ng_g | HORMONE_RESULT_DATA.Raw_ng_g | The "raw" concentration determined in this assay |
| Corrected_ng_g | CORRECTED_HORMONE(HORMONE_RESULT_DATA.Raw_ng_g, HORMONE_KITS.Correction) | The corrected concentration, according to the related HORMONE_KITS.Correction |
| Assay_Date | HORMONE_RESULT_DATA.Assay_Date | Date of this assay |
| Hormone | HORMONE_KITS.Hormone | Code for the hormone whose concentration was assayed |
| Kit | HORMONE_RESULT_DATA.Kit | Code for the kit used in this assay |
| Sample_Comments | HORMONE_SAMPLE_DATA.Comments | Comments about the hormone sample |
| ME_Comments | HORMONE_PREP_DATA.Comments | Comments about the methanol extraction |
| SP_Comments | HORMONE_PREP_DATA.Comments | Comments about the solid-phase extraction |
| Result_Comments | HORMONE_RESULT_DATA.Comments | Comments about the assay |
Contains one row for every sample whose glucocorticoid
concentration has been measured by any kit with a known
correction factor. Results from kits whose HORMONE_KITS.Correction is NULL are
omitted.
Tip
Use this view to see glucocorticoid concentration in all the hormone samples. It joins all the pertinent tables together to gather information, and omits results that are not considered reliable.
This view includes a "Hormone" column that indicates
which hormone was measured in each assay. By definition, this
column will be GC in every
row, so it may seem odd to include the row at all. The column
is retained as a courtesy to users, especially for those who
might unify the rows from this view with rows of other,
similar views (e.g. ESTROGENS, PROGESTERONES, etc.).
Figure 6.26. Query Defining the GLUCOCORTICOIDS View
SELECT hormone_sample_data.tid
, hormone_prep_series.hpsid
, hormone_result_data.hrid
, hormone_sample_data.hsid
, biograph.sname
, tissue_data.collection_date
, tissue_data.collection_date_status AS collection_date_status
, hormone_sample_data.fzdried_date AS fzdried_date
, hormone_sample_data.sifted_date AS sifted_date
, meoh_ext.procedure_date AS me_extracted
, spe.procedure_date AS sp_extracted
, hormone_result_data.raw_ng_g AS raw_ng_g
, corrected_hormone(hormone_result_data.raw_ng_g, hormone_kits.correction) AS corrected_ng_g
, hormone_result_data.assay_date
, hormone_kits.hormone AS hormone
, hormone_result_data.kit AS kit
, hormone_sample_data.comments AS sample_comments
, hormone_result_data.comments AS result_comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ
JOIN hormone_prep_series
ON hormone_prep_series.tid = hormone_sample_data.tid
JOIN hormone_result_data
ON hormone_result_data.hpsid = hormone_prep_series.hpsid
JOIN hormone_kits
ON hormone_kits.kit = hormone_result_data.kit
AND hormone_kits.correction IS NOT NULL
AND hormone_kits.hormone = 'GC'
LEFT JOIN hormone_prep_data AS meoh_ext
ON meoh_ext.procedure = 'MEOH_EXT'
AND meoh_ext.hpsid = hormone_prep_series.hpsid
LEFT JOIN hormone_prep_data AS spe
ON spe.procedure = 'SPE'
AND spe.hpsid = hormone_prep_series.hpsid;
Table 6.12. Columns in the GLUCOCORTICOIDS View
| Column | From | Description |
|---|---|---|
| TId | HORMONE_SAMPLE_DATA.TId | Identifier of the HORMONE_SAMPLE_DATA row and tissue sample |
| HPSId | HORMONE_PREP_SERIES.HPSId | Identifier of the prep series |
| HRId | HORMONE_RESULT_DATA.HRId | Identifier of the assay that generated this result |
| HSId | HORMONE_SAMPLE_DATA.HSId | User-generated identifier for the tissue sample |
| Sname | BIOGRAPH.Sname | Sname of the individual from whom this sample came |
| Collection_Date | TISSUE_DATA.Collection_Date | Date the tissue sample was collected |
| Collection_Date_Status | TISSUE_DATA.Collection_Date_Status | The status of this Collection_Date |
| FzDried_Date | HORMONE_SAMPLE_DATA.FzDried_Date | Date the sample was freeze-dried |
| Sifted_Date | HORMONE_SAMPLE_DATA.Sifted_Date | Date the freeze-dried sample was sifted |
| ME_Extracted | HORMONE_PREP_DATA.Procedure_Date | Date of methanol extraction, in prep for this result |
| SP_Extracted | HORMONE_PREP_DATA.Procedure_Date | Date of solid-phase extraction, in prep for this result |
| Raw_ng_g | HORMONE_RESULT_DATA.Raw_ng_g | The "raw" concentration determined in this assay |
| Corrected_ng_g | CORRECTED_HORMONE(HORMONE_RESULT_DATA.Raw_ng_g, HORMONE_KITS.Correction) | The corrected concentration, according to the related HORMONE_KITS.Correction |
| Assay_Date | HORMONE_RESULT_DATA.Assay_Date | Date of this assay |
| Hormone | HORMONE_KITS.Hormone | Code for the hormone whose concentration was assayed |
| Kit | HORMONE_RESULT_DATA.Kit | Code for the kit used in this assay |
| Sample_Comments | HORMONE_SAMPLE_DATA.Comments | Comments about the hormone sample |
| ME_Comments | HORMONE_PREP_DATA.Comments | Comments about the methanol extraction |
| SP_Comments | HORMONE_PREP_DATA.Comments | Comments about the solid-phase extraction |
| Result_Comments | HORMONE_RESULT_DATA.Comments | Comments about the assay |
Contains one row for every laboratory preparation that was performed on a sample. This view includes columns from BIOGRAPH, HORMONE_PREP_DATA, HORMONE_PREP_SERIES, HORMONE_SAMPLE_DATA, TISSUE_DATA, and UNIQUE_INDIVS in order to portray information in a more user-friendly format. This view is also useful for uploading new data.
Tip
Use this view instead of the HORMONE_PREP_DATA table.
Figure 6.28. Query Defining the HORMONE_PREPS View
SELECT hormone_sample_data.tid AS tid
, hormone_sample_data.hsid AS hsid
, unique_indivs.individ AS individ
, biograph.sname AS sname
, hormone_prep_series.hpsid AS hpsid
, hormone_prep_series.series AS series
, hormone_prep_data.hpid AS hpid
, hormone_prep_data.procedure AS procedure
, hormone_prep_data.procedure_date AS procedure_date
, hormone_prep_data.comments AS comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ
JOIN hormone_prep_series
ON hormone_prep_series.tid = hormone_sample_data.tid
JOIN hormone_prep_data
ON hormone_prep_data.hpsid = hormone_prep_series.hpsid;
Table 6.13. Columns in the HORMONE_PREPS View
| Column | From | Description |
|---|---|---|
| TId | HORMONE_SAMPLE_DATA.TId | Identifier of the HORMONE_SAMPLE_DATA row and tissue sample |
| HSId | HORMONE_SAMPLE_DATA.HSId | User-generated identifier for the tissue sample |
| IndivId | UNIQUE_INDIVS.IndivId | Name/ID for the source individual |
| Sname | BIOGRAPH.Sname | Sname of the source individual |
| HPSId | HORMONE_PREP_SERIES.HPSId | Identifier of the series to which this prep belongs |
| Series | HORMONE_PREP_SERIES.Series | Identifier for the prep series for this sample |
| HPId | HORMONE_PREP_DATA.HPId | Identifier for the HORMONE_PREP_DATA row |
| Procedure | HORMONE_PREP_DATA.Procedure | Code indicating what was done |
| Procedure_Date | HORMONE_PREP_DATA.Procedure_Date | Date that this prep was performed |
| Comments | HORMONE_PREP_DATA.Comments | Miscellaneous notes/comments about this prep |
Inserting a row into HORMONE_PREPS inserts a row in HORMONE_PREP_DATA, as expected. A new HORMONE_PREP_SERIES row may also be inserted, as described below.
When identifying the related tissue sample, either or both of the TId and HSId columns must be provided. If both, they must be related in HORMONE_SAMPLE_DATA.
It is not necessary to provide IndivId or Sname values. Any such values that are provided must match the related values for the provided TId and/or HSId.
If HPSId is provided, it must already be an HPSId value in HORMONE_PREP_SERIES, and its related TId must match the provided TId or be related to the provided HSId.
If a row’s series has not yet been added to HORMONE_PREP_SERIES, this view can add it. When no HPSId is provided, the view will use the provided Series and either TId or HSId values to determine if there is already such a row in HORMONE_PREP_SERIES. If no such HORMONE_PREP_SERIES row is found, then those values are used to create a new HORMONE_PREP_SERIES row. The inserted HORMONE_PREP_DATA.HPSId is either that of the found row or of the newly-created one.
Updating a row in HORMONE_PREPS updates the underlying row in HORMONE_PREP_DATA, as expected.
If both TId and HSId are updated, they must be related in HORMONE_SAMPLE_DATA. If either or both of those columns are updated, either or both of the Series and HPSId columns must also be updated.
To update the HORMONE_PREP_DATA.HPSId column, the HPSId can be updated directly, or the Series can be updated alone. If the Series is updated without the HPSId, this view will use the Series and the TId to look up the correct HPSId from HORMONE_PREP_SERIES. If changing the HPSId also requires a change to the TId and HSId, then an appropriate update to either or both of those columns must be provided at the same time as the update to the HPSId.
If any of the TId, HSId, HPSId, or Series columns
are changed, there must already be a HORMONE_PREP_SERIES row containing the new
values. Unlike on INSERT, this view
will not create a new series in HORMONE_PREP_SERIES on
UPDATE.
Attempts to update the IndivId or Sname columns will return an error.
Tip
To change either of these values, you should update only the TId or HSId column, or update the related TISSUE_DATA row.
Deleting a row from HORMONE_PREPS deletes the underlying row in HORMONE_PREP_DATA, as expected. The related row in HORMONE_PREP_SERIES is unaffected.
Contains one row for every hormone assay result for a sample. That is, every HORMONE_RESULT_DATA row. This view includes columns from BIOGRAPH, HORMONE_PREP_SERIES, HORMONE_RESULT_DATA, HORMONE_SAMPLE_DATA, TISSUE_DATA, and UNIQUE_INDIVS in order to portray information in a more user-friendly format. This view is also useful for uploading new data.
Tip
Use this view instead of the HORMONE_RESULT_DATA table.
Figure 6.30. Query Defining the HORMONE_RESULTS View
SELECT hormone_sample_data.tid AS tid
, hormone_sample_data.hsid AS hsid
, unique_indivs.individ AS individ
, biograph.sname AS sname
, hormone_prep_series.hpsid AS hpsid
, hormone_prep_series.series AS series
, hormone_result_data.hrid AS hrid
, hormone_kits.hormone AS hormone
, hormone_result_data.kit AS kit
, hormone_result_data.assay_date AS assay_date
, hormone_result_data.grams_used AS grams_used
, hormone_result_data.raw_ng_g AS raw_ng_g
, corrected_hormone(hormone_result_data.raw_ng_g, hormone_kits.correction) AS corrected_ng_g
, hormone_result_data.comments AS comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ
JOIN hormone_prep_series
ON hormone_prep_series.tid = hormone_sample_data.tid
JOIN hormone_result_data
ON hormone_result_data.hpsid = hormone_prep_series.hpsid
JOIN hormone_kits
ON hormone_kits.kit = hormone_result_data.kit;
Table 6.14. Columns in the HORMONE_RESULTS View
| Column | From | Description |
|---|---|---|
| TId | HORMONE_SAMPLE_DATA.TId | Identifier of the HORMONE_SAMPLE_DATA row and tissue sample |
| HSId | HORMONE_SAMPLE_DATA.HSId | User-generated identifier for the tissue sample |
| IndivId | UNIQUE_INDIVS.IndivId | Name/ID for the source individual |
| Sname | BIOGRAPH.Sname | Sname of the source individual |
| HPSId | HORMONE_PREP_SERIES.HPSId | Identifier of the series to which this prep belongs |
| Series | HORMONE_PREP_SERIES.Series | Identifier for the prep series for this sample |
| HRId | HORMONE_RESULT_DATA.HRId | Identifier of the HORMONE_RESULT_DATA row |
| Hormone | HORMONE_KITS.Hormone | The hormone that was measured in this result |
| Kit | HORMONE_RESULT_DATA.Kit | The kit used to perform this assay |
| Assay_Date | HORMONE_RESULT_DATA.Assay_Date | The date of this assay |
| Grams_Used | HORMONE_RESULT_DATA.Grams_Used | The mass of tissue in grams that was consumed to generate this result |
| Raw_ng_g | HORMONE_RESULT_DATA.Raw_ng_g | The "raw" (uncorrected) concentration of this hormone in ng/g according to this assay |
| Corrected_ng_g | CORRECTED_HORMONE(HORMONE_RESULT_DATA.Raw_ng_g, HORMONE_KITS.Correction) | The corrected concentration, according to the related HORMONE_KITS.Correction |
| Comments | HORMONE_RESULT_DATA.Comments | Miscellaneous notes/comments about this result |
Inserting a row into HORMONE_RESULTS inserts a row in HORMONE_RESULT_DATA, as expected.
When identifying the related tissue sample, either the TId or HSId must be provided. If both are provided, they must be related in HORMONE_SAMPLE_DATA.
It is not necessary to provide IndivId or Sname values. Any such values that are provided must match the related values for the provided TId or HSId.
When identifying the HORMONE_RESULT_DATA.HPSId to insert, either or both of the HPSId and Series columns must be provided[249].
Any provided HSId, TId, HPSId, and/or Series values must be related in HORMONE_SAMPLE_DATA and HORMONE_PREP_SERIES.
It is not necessary to provide a Hormone value. If one is provided, it must match the related HORMONE_KITS.Hormone for the provided Kit.
It is not necessary to provide a Corrected_ng_g value, as this is a calculated column. If one is provided, it must match the value that is calculated by the corrected_hormone() function with the provided Raw_ng_g and the related HORMONE_KITS.Correction.
Updating a row in HORMONE_RESULTS updates the underlying row in HORMONE_RESULT_DATA, as expected.
If both TId and HSId are updated, they must be related in HORMONE_SAMPLE_DATA. If either or both of those columns are updated, either or both of the Series and HPSId columns must also be updated.
To update the HORMONE_RESULT_DATA.HPSId column, the HPSId can be updated directly, or the Series can be updated alone. If the Series is updated without the HPSId, this view will use the Series and the TId to look up the correct HPSId from HORMONE_PREP_SERIES. If changing the HPSId also requires a change to the TId and HSId, then an appropriate update to either or both of those columns must be provided at the same time as the update to the HPSId.
If any of the TId, HSId, HPSId, or Series columns are changed, there must already be a HORMONE_PREP_SERIES row containing the new values.
Attempts to update the IndivId or Sname columns will return an error.
Tip
To change either of these values, you should update only the TId or HSId column, or update the related TISSUE_DATA row.
Attempts to update the Hormone column will return an error.
Tip
To change this value, you should update the Kit column.
If the Corrected_ng_g is updated, the new value must match the value that is calculated by the corrected_hormone() function with the row's Raw_ng_g and the related HORMONE_KITS.Correction, which will only happen if either or both of the Raw_ng_g and Kit columns is also updated.
Tip
To change the concentration for a row, update the Raw_ng_g and let the system determine the corrected concentration.
Deleting a row from HORMONE_RESULTS deletes the underlying row from HORMONE_RESULT_DATA, as expected.
Contains one row for every tissue sample that has undergone any hormone analysis. That is, for every HORMONE_SAMPLE_DATA row. This view includes columns from BIOGRAPH, HORMONE_SAMPLE_DATA, TISSUE_DATA, and UNIQUE_INDIVS in order to portray information in a more user-friendly format. This view is also useful for uploading new data.
Tip
Use this view instead of the HORMONE_SAMPLE_DATA table.
Figure 6.32. Query Defining the HORMONE_SAMPLES View
SELECT hormone_sample_data.tid AS tid
, hormone_sample_data.hsid AS hsid
, unique_indivs.individ AS individ
, biograph.sname AS sname
, tissue_data.collection_date AS collection_date
, tissue_data.collection_date_status AS collection_date_status
, hormone_sample_data.fzdried_date AS fzdried_date
, hormone_sample_data.sifted_date AS sifted_date
, hormone_sample_data.avail_mass_g AS avail_mass_g
, hormone_sample_data.avail_date AS avail_date
, hormone_sample_data.comments AS comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ;
Table 6.15. Columns in the HORMONE_SAMPLES View
| Column | From | Description |
|---|---|---|
| TId | HORMONE_SAMPLE_DATA.TId | Identifier of the HORMONE_SAMPLE_DATA row and tissue sample |
| HSId | HORMONE_SAMPLE_DATA.HSId | User-generated identifier for the tissue sample |
| IndivId | UNIQUE_INDIVS.IndivId | Name/ID for the source individual |
| Sname | BIOGRAPH.Sname | Sname of the source individual |
| Collection_Date | TISSUE_DATA.Collection_Date | Date the tissue sample was collected |
| Collection_Date_Status | TISSUE_DATA.Collection_Date_Status | The status of this Collection_Date |
| FzDried_Date | HORMONE_SAMPLE_DATA.FzDried_Date | Date the sample was freeze-dried |
| Sifted_Date | HORMONE_SAMPLE_DATA.Sifted_Date | Date the freeze-dried sample was sifted |
| Avail_Mass_g | HORMONE_SAMPLE_DATA.Avail_Mass_g | Amount of sample (in g) remaining in the tube, as of the Avail_Date |
| Avail_Date | HORMONE_SAMPLE_DATA.Avail_Date | Date that the Avail_Mass_g was determined |
| Comments | HORMONE_SAMPLE_DATA.Comments | Miscellaneous notes/comments about this sample |
Inserting a row into HORMONE_SAMPLES inserts a row in HORMONE_SAMPLE_DATA, as expected.
It is not necessary to provide IndivId, Sname, or Collection_Date values. Any such values that are provided must match the related values for the provided TId.
Updating a row in HORMONE_SAMPLES updates the underlying row in HORMONE_SAMPLE_DATA, as expected.
Attempts to update the IndivId, Sname, or Collection_Date columns will return an error.
Tip
To change any of these values for a sample, update the related TISSUE_DATA row.
Deleting a row from HORMONE_SAMPLES deletes the underlying row in HORMONE_SAMPLE_DATA, as expected. The related row in TISSUE_DATA is unaffected.
This view shows all the hybrid morphology scores, one line per report.
Figure 6.34. Query Defining the HYBRIDMORPH_SCORES View
WITH concat_observers AS (SELECT hmrid
, string_agg(observer, '/' ORDER BY hmoid) AS observers
FROM hybridmorph_observers
GROUP BY hmrid)
SELECT hybridmorph_reports.hmrid AS hmrid
, hybridmorph_reports.sname AS sname
, hybridmorph_reports.date AS date
, hybridmorph_reports.grp AS grp
, concat_observers.observers AS observers
, color.score AS color
, hair_leng.score AS hair_leng
, body_shape.score AS body_shape
, head_shape.score AS head_shape
, tail_leng.score AS tail_leng
, tail_bend.score AS tail_bend
, muzzle.score AS muzzle
FROM hybridmorph_reports
LEFT JOIN concat_observers
ON concat_observers.hmrid = hybridmorph_reports.hmrid
LEFT JOIN hybridmorph_score_data AS color
ON color.hmrid = hybridmorph_reports.hmrid
AND color.trait = 'COLOR'
LEFT JOIN hybridmorph_score_data AS hair_leng
ON hair_leng.hmrid = hybridmorph_reports.hmrid
AND hair_leng.trait = 'HAIR_LENG'
LEFT JOIN hybridmorph_score_data AS body_shape
ON body_shape.hmrid = hybridmorph_reports.hmrid
AND body_shape.trait = 'BODY_SHAPE'
LEFT JOIN hybridmorph_score_data AS head_shape
ON head_shape.hmrid = hybridmorph_reports.hmrid
AND head_shape.trait = 'HEAD_SHAPE'
LEFT JOIN hybridmorph_score_data AS tail_leng
ON tail_leng.hmrid = hybridmorph_reports.hmrid
AND tail_leng.trait = 'TAIL_LENG'
LEFT JOIN hybridmorph_score_data AS tail_bend
ON tail_bend.hmrid = hybridmorph_reports.hmrid
AND tail_bend.trait = 'TAIL_BEND'
LEFT JOIN hybridmorph_score_data AS muzzle
ON muzzle.hmrid = hybridmorph_reports.hmrid
AND muzzle.trait = 'MUZZLE';
Table 6.16. Columns in the HYBRIDMORPH_SCORES View
| Column | From | Description |
|---|---|---|
| HMRId | HYBRIDMORPH_REPORTS.HMRId | Identifier for the report. |
| Sname | HYBRIDMORPH_REPORTS.Sname | The scored individual. |
| Date | HYBRIDMORPH_REPORTS.Date | The date of the scoring. |
| Grp | HYBRIDMORPH_REPORTS.Grp | The individual's group on this date, according to the observer(s). |
| Observers | HYBRIDMORPH_OBSERVERS.Observer | All of this report's observers, concatenated
together and separated by a
"/". If no
related observers, then NULL. |
| Color | HYBRIDMORPH_SCORE_DATA.Score | The individual's "Coat Color" score. |
| Hair_Leng | HYBRIDMORPH_SCORE_DATA.Score | The individual's "Hair Length" score. |
| Body_Shape | HYBRIDMORPH_SCORE_DATA.Score | The individual's "Body Shape" score. |
| Head_Shape | HYBRIDMORPH_SCORE_DATA.Score | The individual's "Head Shape" score. |
| Tail_Leng | HYBRIDMORPH_SCORE_DATA.Score | The individual's "Tail Length" score. |
| Tail_Bend | HYBRIDMORPH_SCORE_DATA.Score | The individual's "Tail_Bend" score. |
| Muzzle | HYBRIDMORPH_SCORE_DATA.Score | The individual's "Muzzle" score. |
Inserting a row into this view inserts rows into
HYBRIDMORPH_REPORTS, HYBRIDMORPH_OBSERVERS, and HYBRIDMORPH_SCORE_DATA as expected. If one of
the "score" columns is NULL, a HYBRIDMORPH_SCORE_DATA row with that column's
Trait is NOT added.
Updating a row in this view updates rows in HYBRIDMORPH_REPORTS, HYBRIDMORPH_OBSERVERS, and HYBRIDMORPH_SCORE_DATA as expected.
The HMRId cannot be changed.
Deleting a row from this view deletes from HYBRIDMORPH_REPORTS, HYBRIDMORPH_OBSERVERS, and HYBRIDMORPH_SCORE_DATA as expected.
Contains one row for every sample whose progesterone
concentration has been measured by any kit with a known
correction factor. Results from kits whose HORMONE_KITS.Correction is NULL are
omitted.
Tip
Use this view to see progesterone concentration in all the hormone samples. It joins all the pertinent tables together to gather information, and omits results that are not considered reliable.
This view includes a "Hormone" column that indicates
which hormone was measured in each assay. By definition, this
column will be P in every
row, so it may seem odd to include the row at all. The column
is retained as a courtesy to users, especially for those who
might unify the rows from this view with rows of other,
similar views (e.g. ESTROGENS, GLUCOCORTICOIDS, etc.).
Figure 6.36. Query Defining the PROGESTERONES View
SELECT hormone_sample_data.tid
, hormone_prep_series.hpsid
, hormone_result_data.hrid
, hormone_sample_data.hsid
, biograph.sname
, tissue_data.collection_date
, tissue_data.collection_date_status AS collection_date_status
, hormone_sample_data.fzdried_date AS fzdried_date
, hormone_sample_data.sifted_date AS sifted_date
, meoh_ext.procedure_date AS me_extracted
, spe.procedure_date AS sp_extracted
, hormone_result_data.raw_ng_g AS raw_ng_g
, corrected_hormone(hormone_result_data.raw_ng_g, hormone_kits.correction) AS corrected_ng_g
, hormone_result_data.assay_date
, hormone_kits.hormone AS hormone
, hormone_result_data.kit AS kit
, hormone_sample_data.comments AS sample_comments
, hormone_result_data.comments AS result_comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ
JOIN hormone_prep_series
ON hormone_prep_series.tid = hormone_sample_data.tid
JOIN hormone_result_data
ON hormone_result_data.hpsid = hormone_prep_series.hpsid
JOIN hormone_kits
ON hormone_kits.kit = hormone_result_data.kit
AND hormone_kits.correction IS NOT NULL
AND hormone_kits.hormone = 'P'
LEFT JOIN hormone_prep_data AS meoh_ext
ON meoh_ext.procedure = 'MEOH_EXT'
AND meoh_ext.hpsid = hormone_prep_series.hpsid
LEFT JOIN hormone_prep_data AS spe
ON spe.procedure = 'SPE'
AND spe.hpsid = hormone_prep_series.hpsid;
Table 6.17. Columns in the PROGESTERONES View
| Column | From | Description |
|---|---|---|
| TId | HORMONE_SAMPLE_DATA.TId | Identifier of the HORMONE_SAMPLE_DATA row and tissue sample |
| HPSId | HORMONE_PREP_SERIES.HPSId | Identifier of the prep series |
| HRId | HORMONE_RESULT_DATA.HRId | Identifier of the assay that generated this result |
| HSId | HORMONE_SAMPLE_DATA.HSId | User-generated identifier for the tissue sample |
| Sname | BIOGRAPH.Sname | Sname of the individual from whom this sample came |
| Collection_Date | TISSUE_DATA.Collection_Date | Date the tissue sample was collected |
| Collection_Date_Status | TISSUE_DATA.Collection_Date_Status | The status of this Collection_Date |
| FzDried_Date | HORMONE_SAMPLE_DATA.FzDried_Date | Date the sample was freeze-dried |
| Sifted_Date | HORMONE_SAMPLE_DATA.Sifted_Date | Date the freeze-dried sample was sifted |
| ME_Extracted | HORMONE_PREP_DATA.Procedure_Date | Date of methanol extraction, in prep for this result |
| SP_Extracted | HORMONE_PREP_DATA.Procedure_Date | Date of solid-phase extraction, in prep for this result |
| Raw_ng_g | HORMONE_RESULT_DATA.Raw_ng_g | The "raw" concentration determined in this assay |
| Corrected_ng_g | CORRECTED_HORMONE(HORMONE_RESULT_DATA.Raw_ng_g, HORMONE_KITS.Correction) | The corrected concentration, according to the related HORMONE_KITS.Correction |
| Assay_Date | HORMONE_RESULT_DATA.Assay_Date | Date of this assay |
| Hormone | HORMONE_KITS.Hormone | Code for the hormone whose concentration was assayed |
| Kit | HORMONE_RESULT_DATA.Kit | Code for the kit used in this assay |
| Sample_Comments | HORMONE_SAMPLE_DATA.Comments | Comments about the hormone sample |
| ME_Comments | HORMONE_PREP_DATA.Comments | Comments about the methanol extraction |
| SP_Comments | HORMONE_PREP_DATA.Comments | Comments about the solid-phase extraction |
| Result_Comments | HORMONE_RESULT_DATA.Comments | Comments about the assay |
Contains one row for every sample whose testosterone
concentration has been measured by any kit with a known
correction factor. Results from kits whose HORMONE_KITS.Correction is NULL are
omitted.
Tip
Use this view to see testosterone concentration in all the hormone samples. It joins all the pertinent tables together to gather information, and omits results that are not considered reliable.
This view includes a "Hormone" column that indicates
which hormone was measured in each assay. By definition, this
column will be T in every
row, so it may seem odd to include the row at all. The column
is retained as a courtesy to users, especially for those who
might unify the rows from this view with rows of other,
similar views (e.g. ESTROGENS, GLUCOCORTICOIDS, etc.).
Figure 6.38. Query Defining the TESTOSTERONES View
SELECT hormone_sample_data.tid
, hormone_prep_series.hpsid
, hormone_result_data.hrid
, hormone_sample_data.hsid
, biograph.sname
, tissue_data.collection_date
, tissue_data.collection_date_status AS collection_date_status
, hormone_sample_data.fzdried_date AS fzdried_date
, hormone_sample_data.sifted_date AS sifted_date
, meoh_ext.procedure_date AS me_extracted
, spe.procedure_date AS sp_extracted
, hormone_result_data.raw_ng_g AS raw_ng_g
, corrected_hormone(hormone_result_data.raw_ng_g, hormone_kits.correction) AS corrected_ng_g
, hormone_result_data.assay_date
, hormone_kits.hormone AS hormone
, hormone_result_data.kit AS kit
, hormone_sample_data.comments AS sample_comments
, hormone_result_data.comments AS result_comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ
JOIN hormone_prep_series
ON hormone_prep_series.tid = hormone_sample_data.tid
JOIN hormone_result_data
ON hormone_result_data.hpsid = hormone_prep_series.hpsid
JOIN hormone_kits
ON hormone_kits.kit = hormone_result_data.kit
AND hormone_kits.correction IS NOT NULL
AND hormone_kits.hormone = 'T'
LEFT JOIN hormone_prep_data AS meoh_ext
ON meoh_ext.procedure = 'MEOH_EXT'
AND meoh_ext.hpsid = hormone_prep_series.hpsid
LEFT JOIN hormone_prep_data AS spe
ON spe.procedure = 'SPE'
AND spe.hpsid = hormone_prep_series.hpsid;
Table 6.18. Columns in the TESTOSTERONES View
| Column | From | Description |
|---|---|---|
| TId | HORMONE_SAMPLE_DATA.TId | Identifier of the HORMONE_SAMPLE_DATA row and tissue sample |
| HPSId | HORMONE_PREP_SERIES.HPSId | Identifier of the prep series |
| HRId | HORMONE_RESULT_DATA.HRId | Identifier of the assay that generated this result |
| HSId | HORMONE_SAMPLE_DATA.HSId | User-generated identifier for the tissue sample |
| Sname | BIOGRAPH.Sname | Sname of the individual from whom this sample came |
| Collection_Date | TISSUE_DATA.Collection_Date | Date the tissue sample was collected |
| Collection_Date_Status | TISSUE_DATA.Collection_Date_Status | The status of this Collection_Date |
| FzDried_Date | HORMONE_SAMPLE_DATA.FzDried_Date | Date the sample was freeze-dried |
| Sifted_Date | HORMONE_SAMPLE_DATA.Sifted_Date | Date the freeze-dried sample was sifted |
| ME_Extracted | HORMONE_PREP_DATA.Procedure_Date | Date of methanol extraction, in prep for this result |
| SP_Extracted | HORMONE_PREP_DATA.Procedure_Date | Date of solid-phase extraction, in prep for this result |
| Raw_ng_g | HORMONE_RESULT_DATA.Raw_ng_g | The "raw" concentration determined in this assay |
| Corrected_ng_g | CORRECTED_HORMONE(HORMONE_RESULT_DATA.Raw_ng_g, HORMONE_KITS.Correction) | The corrected concentration, according to the related HORMONE_KITS.Correction |
| Assay_Date | HORMONE_RESULT_DATA.Assay_Date | Date of this assay |
| Hormone | HORMONE_KITS.Hormone | Code for the hormone whose concentration was assayed |
| Kit | HORMONE_RESULT_DATA.Kit | Code for the kit used in this assay |
| Sample_Comments | HORMONE_SAMPLE_DATA.Comments | Comments about the hormone sample |
| ME_Comments | HORMONE_PREP_DATA.Comments | Comments about the methanol extraction |
| SP_Comments | HORMONE_PREP_DATA.Comments | Comments about the solid-phase extraction |
| Result_Comments | HORMONE_RESULT_DATA.Comments | Comments about the assay |
Contains one row for every sample whose thyroid hormone
concentration has been measured by any kit with a known
correction factor. Results from kits whose HORMONE_KITS.Correction is NULL are
omitted.
Tip
Use this view to see thyroid hormone concentration in all the hormone samples. It joins all the pertinent tables together to gather information, and omits results that are not considered reliable.
This view includes a "Hormone" column that indicates
which hormone was measured in each assay. By definition, this
column will be TH in every
row, so it may seem odd to include the row at all. The column
is retained as a courtesy to users, especially for those who
might unify the rows from this view with rows of other,
similar views (e.g. ESTROGENS, GLUCOCORTICOIDS, etc.).
Figure 6.40. Query Defining the THYROID_HORMONES View
SELECT hormone_sample_data.tid
, hormone_prep_series.hpsid
, hormone_result_data.hrid
, hormone_sample_data.hsid
, biograph.sname
, tissue_data.collection_date
, tissue_data.collection_date_status AS collection_date_status
, hormone_sample_data.fzdried_date AS fzdried_date
, hormone_sample_data.sifted_date AS sifted_date
, etoh_ext.procedure_date AS et_extracted
, hormone_result_data.raw_ng_g AS raw_ng_g
, corrected_hormone(hormone_result_data.raw_ng_g, hormone_kits.correction) AS corrected_ng_g
, hormone_result_data.assay_date
, hormone_kits.hormone AS hormone
, hormone_result_data.kit AS kit
, hormone_sample_data.comments AS sample_comments
, hormone_result_data.comments AS result_comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ
JOIN hormone_prep_series
ON hormone_prep_series.tid = hormone_sample_data.tid
JOIN hormone_result_data
ON hormone_result_data.hpsid = hormone_prep_series.hpsid
JOIN hormone_kits
ON hormone_kits.kit = hormone_result_data.kit
AND hormone_kits.correction IS NOT NULL
AND hormone_kits.hormone = 'TH'
LEFT JOIN hormone_prep_data AS etoh_ext
ON etoh_ext.procedure = 'ETOH_EXT'
AND etoh_ext.hpsid = hormone_prep_series.hpsid;
Table 6.19. Columns in the THYROID_HORMONES View
| Column | From | Description |
|---|---|---|
| TId | HORMONE_SAMPLE_DATA.TId | Identifier of the HORMONE_SAMPLE_DATA row and tissue sample |
| HPSId | HORMONE_PREP_SERIES.HPSId | Identifier of the prep series |
| HRId | HORMONE_RESULT_DATA.HRId | Identifier of the assay that generated this result |
| HSId | HORMONE_SAMPLE_DATA.HSId | User-generated identifier for the tissue sample |
| Sname | BIOGRAPH.Sname | Sname of the individual from whom this sample came |
| Collection_Date | TISSUE_DATA.Collection_Date | Date the tissue sample was collected |
| Collection_Date_Status | TISSUE_DATA.Collection_Date_Status | The status of this Collection_Date |
| FzDried_Date | HORMONE_SAMPLE_DATA.FzDried_Date | Date the sample was freeze-dried |
| Sifted_Date | HORMONE_SAMPLE_DATA.Sifted_Date | Date the freeze-dried sample was sifted |
| ET_Extracted | HORMONE_PREP_DATA.Procedure_Date | Date of ethanol extraction, in prep for this result |
| Raw_ng_g | HORMONE_RESULT_DATA.Raw_ng_g | The "raw" concentration determined in this assay |
| Corrected_ng_g | CORRECTED_HORMONE(HORMONE_RESULT_DATA.Raw_ng_g, HORMONE_KITS.Correction) | The corrected concentration, according to the related HORMONE_KITS.Correction |
| Assay_Date | HORMONE_RESULT_DATA.Assay_Date | Date of this assay |
| Hormone | HORMONE_KITS.Hormone | Code for the hormone whose concentration was assayed |
| Kit | HORMONE_RESULT_DATA.Kit | Code for the kit used in this assay |
| Sample_Comments | HORMONE_SAMPLE_DATA.Comments | Comments about the hormone sample |
| EE_Comments | HORMONE_PREP_DATA.Comments | Comments about the ethanol extraction |
| Result_Comments | HORMONE_RESULT_DATA.Comments | Comments about the assay |
This view is intended to be the main place to visualize the wounds/pathologies data without heal updates. It contains one row for every body part affected in a wound/pathology cluster, including all related columns from WP_REPORTS, WP_DETAILS, WP_AFFECTEDPARTS and BODYPARTS, and a single column of concatenated observers from WP_OBSERVERS.
Tip
Use this view instead of the individual component tables when selecting wounds/pathologies data and when heal updates needn't be included.
Figure 6.42. Query Defining the WOUNDSPATHOLOGIES View
WITH concat_observers AS (SELECT wprid
, string_agg(observer, '/' ORDER BY wpoid) as observers
FROM wp_observers
GROUP BY wprid)
SELECT wp_reports.wprid AS wprid
, wp_reports.wid AS wid
, wp_reports.date AS reportdate
, wp_reports.time AS reporttime
, concat_observers.observers AS observers
, wp_reports.sname AS sname
, wp_reports.grp AS grp
, wp_reports.observercomments AS observercomments
, wp_reports.reportstate AS reportstate
, wp_details.wpdid AS wpdid
, wp_details.woundpathcode AS woundpathcode
, wp_details.cluster AS cluster
, wp_details.maxdimension AS maxdimension
, wp_details.impairslocomotion AS impairslocomotion
, wp_details.infectionsigns AS infectionsigns
, wp_details.notes AS detailnotes
, wp_affectedparts.wpaid AS wpaid
, wp_affectedparts.bodypart AS bodypart
, bodyparts.bodyside AS bodyside
, bodyparts.innerouter AS innerouter
, bodyparts.bodyregion AS bodyregion
, wp_affectedparts.quantity_affecting_part AS quantity_affecting_part
FROM wp_reports
LEFT JOIN concat_observers
ON concat_observers.wprid = wp_reports.wprid
LEFT JOIN wp_details
ON wp_details.wprid = wp_reports.wprid
LEFT JOIN wp_affectedparts
ON wp_affectedparts.wpdid = wp_details.wpdid
LEFT JOIN bodyparts
ON bodyparts.bpid = wp_affectedparts.bodypart;
Table 6.20. Columns in the WOUNDSPATHOLOGIES View
| Column | From | Description |
|---|---|---|
| WPRId | WP_REPORTS.WPRId | Identifier for the report. |
| WId | WP_REPORTS.WId | User-generated identifier for the report. |
| ReportDate | WP_REPORTS.Date | The date that the report was created. |
| ReportTime | WP_REPORTS.Time | The time that the report was created. |
| Observers | WP_OBSERVERS.Observer | All of the report's observers, concatenated
together and separated by a
"/". If no
observers, then NULL. |
| Sname | WP_REPORTS.Sname | The sname of the affected individual. |
| Grp | WP_REPORTS.Grp | The group of the individual in the report, according to the observer(s). |
| ObserverComments | WP_REPORTS.ObserverComments | Notes or comments from the observer(s) about the report. |
| ReportState | WP_REPORTS.ReportState | Status of the report. |
| WPDId | WP_DETAILS.WPDId | Identifier for the wound/pathology cluster. |
| WoundPathCode | WP_DETAILS.WoundPathCode | Code indicating the wound/pathology type for the cluster. |
| Cluster | WP_DETAILS.Cluster | The wound/pathology cluster identifier. |
| MaxDimension | WP_DETAILS.MaxDimension | The highest observed length, height, depth, etc. (as applicable), in cm, of all wounds/pathologies in the cluster. |
| ImpairsLocomotion | WP_DETAILS.ImpairsLocomotion | Boolean indicating whether or not the wound/pathology cluster impairs the individual's locomotion. |
| InfectionSigns | WP_DETAILS.InfectionSigns | Boolean indicating whether or not the wound/pathology cluster includes signs of an infection. |
| DetailNotes | WP_DETAILS.Notes | Textual comments or notes about the cluster. |
| WPAId | WP_AFFECTEDPARTS.WPAId | Identifier for the affected body part in the wound/pathology cluster. |
| Bodypart | WP_AFFECTEDPARTS.Bodypart | Unique identifier for the body part. |
| Bodyside | BODYPARTS.Bodyside | Code indicating the side of the body on which the affected part is located. |
| Innerouter | BODYPARTS.Innerouter | Code indicating if the affected body part is on the inner or outer side of the body part. |
| Bodyregion | BODYPARTS.Bodyregion | Code indicating the region on the body of the affected body part. |
| Quantity_Affecting_Part | WP_AFFECTEDPARTS.Quantity_Affecting_Part | The number of wounds/pathologies described in the cluster affecting this body part. |
Contains one row for every row in WP_AFFECTEDPARTS, with related identifier columns from WP_REPORTS and related data from the WP_DETAILS and BODYPARTS tables.
The intended purpose of this view is for uploading data into WP_DETAILS and WP_AFFECTEDPARTS. It may also be useful for querying/accessing the data.
Figure 6.44. Query Defining the WP_DETAILS_AFFECTEDPARTS View
SELECT wp_details.wpdid AS wpdid
, wp_reports.wprid AS wprid
, wp_reports.wid AS wid
, wp_details.woundpathcode AS woundpathcode
, wp_details.cluster AS cluster
, wp_details.maxdimension AS maxdimension
, wp_details.impairslocomotion AS impairslocomotion
, wp_details.infectionsigns AS infectionsigns
, wp_details.notes AS detailnotes
, wp_affectedparts.wpaid AS wpaid
, wp_affectedparts.wpdid AS bodypart_wpdid
, wp_affectedparts.bodypart AS bodypart
, bodyparts.bodyside AS bodyside
, bodyparts.innerouter AS innerouter
, bodyparts.bodyregion AS bodyregion
, wp_affectedparts.quantity_affecting_part AS quantity_affecting_part
FROM wp_reports
JOIN wp_details
ON wp_details.wprid = wp_reports.wprid
LEFT JOIN wp_affectedparts
ON wp_affectedparts.wpdid = wp_details.wpdid
LEFT JOIN bodyparts
ON bodyparts.bpid = wp_affectedparts.bodypart;
Table 6.21. Columns in the WP_DETAILS_AFFECTEDPARTS View
| Column | From | Description |
|---|---|---|
| WPDId | WP_DETAILS.WPDId | Identifier for the wound/pathology cluster. |
| WPRId | WP_REPORTS.WPRId | Identifier for the report in which these wounds/pathologies were recorded. |
| WId | WP_REPORTS.WId | User-generated identifier for the report in which these wounds/pathologies were recorded. |
| WoundPathCode | WP_DETAILS.WoundPathCode | Code indicating the wound/pathology type. |
| Cluster | WP_DETAILS.Cluster | The wound/pathology cluster identifier. |
| MaxDimension | WP_DETAILS.MaxDimension | The highest observed length, height, depth, etc. (as applicable), in cm, of all wounds/pathologies in this cluster. |
| ImpairsLocomotion | WP_DETAILS.ImpairsLocomotion | Boolean indicating whether or not this wound/pathology cluster impairs the individual's locomotion. |
| InfectionSigns | WP_DETAILS.InfectionSigns | Boolean indicating whether or not this wound/pathology cluster includes signs of an infection. |
| DetailNotes | WP_DETAILS.Notes | Textual comments or notes about this cluster. |
| WPAId | WP_AFFECTEDPARTS.WPAId | Identifier for the affected body part in this
wound/pathology cluster. If there are no related
rows in WP_AFFECTEDPARTS, then
NULL. |
| Bodypart_WPDId | WP_AFFECTEDPARTS.WPDId | Identifier for the wound/pathology cluster,
from WP_AFFECTEDPARTS. When
selecting data, this will always equal the WPDId
column. This column is included to allow the ability
to change the WP_AFFECTEDPARTS.WPDId with an
UPDATE command. If there are no
related rows in WP_AFFECTEDPARTS,
then NULL. |
| Bodypart | WP_AFFECTEDPARTS.Bodypart, BODYPARTS.Bpid | Unique identifier for the body part. If there
are no related rows in WP_AFFECTEDPARTS, then NULL. |
| Bodyside | BODYPARTS.Bodyside | Code indicating the side of the body on which
the affected part is located. If there are no
related rows in WP_AFFECTEDPARTS,
then NULL. |
| Innerouter | BODYPARTS.Innerouter | Code indicating if the affected body part is
on the inner or outer side of the body part. If
there are no related rows in WP_AFFECTEDPARTS, then NULL. |
| Bodyregion | BODYPARTS.Bodyregion | Code indicating the region on the body of the
affected body part. If there are no related rows in
WP_AFFECTEDPARTS, then
NULL. |
| Quantity_Affecting_Part | WP_AFFECTEDPARTS.Quantity_Affecting_Part | The number of wounds/pathologies described in
the cluster affecting this body part. If there are
no related rows in WP_AFFECTEDPARTS, then NULL. |
At least one of the WPRId and WId columns cannot be
NULL; these values are used to determine the related WP_DETAILS.WPRId. If both are provided, that
pair must already exist as a WPRId-WId pair in WP_REPORTS.
There
must be enough body part information provided to identify a
single body part code for the WP_AFFECTEDPARTS.Bodypart column. This means
that the provided Bodypart column must not be NULL or the
provided Bodyside-Innerouter-Bodyregion must be identical to
those of a single row in BODYPARTS. If
the Bodypart is not NULL and one or more of the Bodyside,
Innerouter, or Bodyregion columns is also not NULL, it is
an error if any of the provided Bodyside, Innerouter, or
Bodyregion values does not match their related columns in
BODYPARTS for the provided Bodypart (Bpid).
Inserting a row into WP_DETAILS_AFFECTEDPARTS inserts a row into WP_DETAILS and WP_AFFECTEDPARTS, as described below.
Like their related columns in WP_DETAILS, the WoundPathCode, Cluster,
ImpairsLocomotion, and InfectionSigns columns cannot be
NULL. When there is already a WP_DETAILS row with the provided
WoundPathCode, Cluster, MaxDimension, ImpairsLocomotion,
InfectionSigns, DetailNotes (Notes), and either WPRId or related
WId, a new WP_DETAILS row is not added.
These values are still used to determine the correct WPDId
to use when inserting data into WP_AFFECTEDPARTS.
When Bodypart_WPDId is not provided, new WP_AFFECTEDPARTS rows are inserted using the WPDId of the related WP_DETAILS row. If a Bodypart_WPDId is provided, it must equal the related WPDId from WP_DETAILS, whether or not WPDId is provided.
The new WP_AFFECTEDPARTS.Bodypart value is determined
with the provided body part columns, as described above.
When the Bodypart column is NULL, it is an error if one
or more of the Bodyside, Innerouter, or Bodyregion columns
is also NULL.
Updating a row in WP_DETAILS_AFFECTEDPARTS updates the underlying columns in WP_DETAILS and WP_AFFECTEDPARTS, as expected.
Tip
To update the WPDId in a WP_AFFECTEDPARTS row, update the Bodypart_WPDId column, not the WPDId. The former exists explicitly for this purpose, while the latter refers to the WPDId column in WP_DETAILS, which cannot be changed.
Deleting a row in WP_DETAILS_AFFECTEDPARTS deletes the underlying rows in WP_DETAILS and in WP_AFFECTEDPARTS.
DELETE commands in this view
remove the WP_DETAILS row, and all
related WP_AFFECTEDPARTS rows are
deleted concomitantly. It is not possible to
only remove row(s) from WP_AFFECTEDPARTS when deleting from this
view.
Tip
To remove WP_AFFECTEDPARTS rows without deleting the related WP_DETAILS row, don't use this view. You should manually delete the rows from the WP_AFFECTEDPARTS table.
Contains one row for every row in WP_HEALUPDATES, with related columns from WP_REPORTS, WP_DETAILS, WP_AFFECTEDPARTS, and BODYPARTS, and an "Observers" column concatenating all related WP_OBSERVERS.Observer values together. Whether or not a particular table's rows are "related" depends somewhat on the specificity of the heal update, as discussed below.
Although the relationship between WP_REPORTS, WP_OBSERVERS, WP_DETAILS, WP_AFFECTEDPARTS,
and BODYPARTS rows is unambiguous, the
relationship between them and a particular heal update may not
be so straightforward. For example, a heal update for a
particular cluster (when the WP_HEALUPDATES.WPDId is not NULL) might in
reality apply to one or all of that cluster's affected body
parts, but the update's being recorded for the cluster
indicates that it is unspecified or unknown which specific
body parts have healed. Users may decide on their own to make
assumptions about which body parts are included in such an
update, but it would be misleading for this view to join them
together and imply more specificity than is actually known.
To prevent such false implications of specificity, this view
leaves NULL any columns that are more specific than what is
indicated in the WP_HEALUPDATES
row. Specifically: when the update is for a report (the WPRId is not NULL), the related
values from WP_REPORTS will be returned,
while those from WP_DETAILS, WP_AFFECTEDPARTS, and BODYPARTS
will be NULL. When the update is for a cluster (the WPDId is not NULL), the related
values from WP_REPORTS and WP_DETAILS will be returned, but those from WP_AFFECTEDPARTS and BODYPARTS
will be NULL. An update for an affected body part (the WPAId is not NULL) will return
the related values from all the aforementioned tables.
Regardless of update specificity, the concatenated
"Observers" column will always be included. It will be NULL
only when there are no observers recorded for the related
report in WP_OBSERVERS.
Caution
Many of the tables in this view have a "one-to-many" relationship with each other. Because of this, some normally-unique values may appear to be duplicated across multiple rows. Remember, only WP_HEALS rows are truly unique in this view.
Tip
Use this view when selecting wounds/pathologies data that include the heal updates. (Use this instead of the WP_HEALUPDATES table.) This view presents the data in a format more hospitable for humans to read, and performs the somewhat-tricky task of joining the different ID columns (WPRId, WPDId, and WPAId) to their respective tables.
Figure 6.46. Query Defining the WP_HEALS View
WITH concat_observers AS (SELECT wprid
, string_agg(observer, '/' ORDER BY wpoid) as observers
FROM wp_observers
GROUP BY wprid)
SELECT wp_reports.wprid AS wprid
, wp_reports.wid AS wid
, wp_reports.date AS reportdate
, wp_reports.time AS reporttime
, concat_observers.observers AS observers
, wp_reports.sname AS sname
, wp_reports.grp AS grp
, wp_reports.observercomments AS observercomments
, wp_reports.reportstate AS reportstate
, wp_details.wpdid AS wpdid
, wp_details.woundpathcode AS woundpathcode
, wp_details.cluster AS cluster
, wp_details.maxdimension AS maxdimension
, wp_details.impairslocomotion AS impairslocomotion
, wp_details.infectionsigns AS infectionsigns
, wp_details.notes AS detailnotes
, wp_affectedparts.wpaid AS wpaid
, wp_affectedparts.bodypart AS bodypart
, bodyparts.bodyside AS bodyside
, bodyparts.innerouter AS innerouter
, bodyparts.bodyregion AS bodyregion
, wp_affectedparts.quantity_affecting_part AS quantity_affecting_part
, wp_healupdates.wphid AS wphid
, wp_healupdates.date AS healdate
, wp_healupdates.healstatus AS healstatus
, wp_healupdates.notes AS healnotes
FROM wp_healupdates
LEFT JOIN wp_affectedparts
ON wp_affectedparts.wpaid = wp_healupdates.wpaid
LEFT JOIN bodyparts
ON bodyparts.bpid = wp_affectedparts.bodypart
LEFT JOIN wp_details
ON wp_details.wpdid = COALESCE(wp_affectedparts.wpdid, wp_healupdates.wpdid)
LEFT JOIN wp_reports
ON wp_reports.wprid = COALESCE(wp_details.wprid, wp_healupdates.wprid)
LEFT JOIN concat_observers
ON concat_observers.wprid = wp_reports.wprid;
Figure 6.48. Entity Relationship Diagram of the WP_HEALS View for rows with an update to a wound/pathology report
 |
Figure 6.49. Entity Relationship Diagram of the WP_HEALS View for rows with an update to a wound/pathology cluster
 |
Figure 6.50. Entity Relationship Diagram of the WP_HEALS View for rows with an update to an affected body part
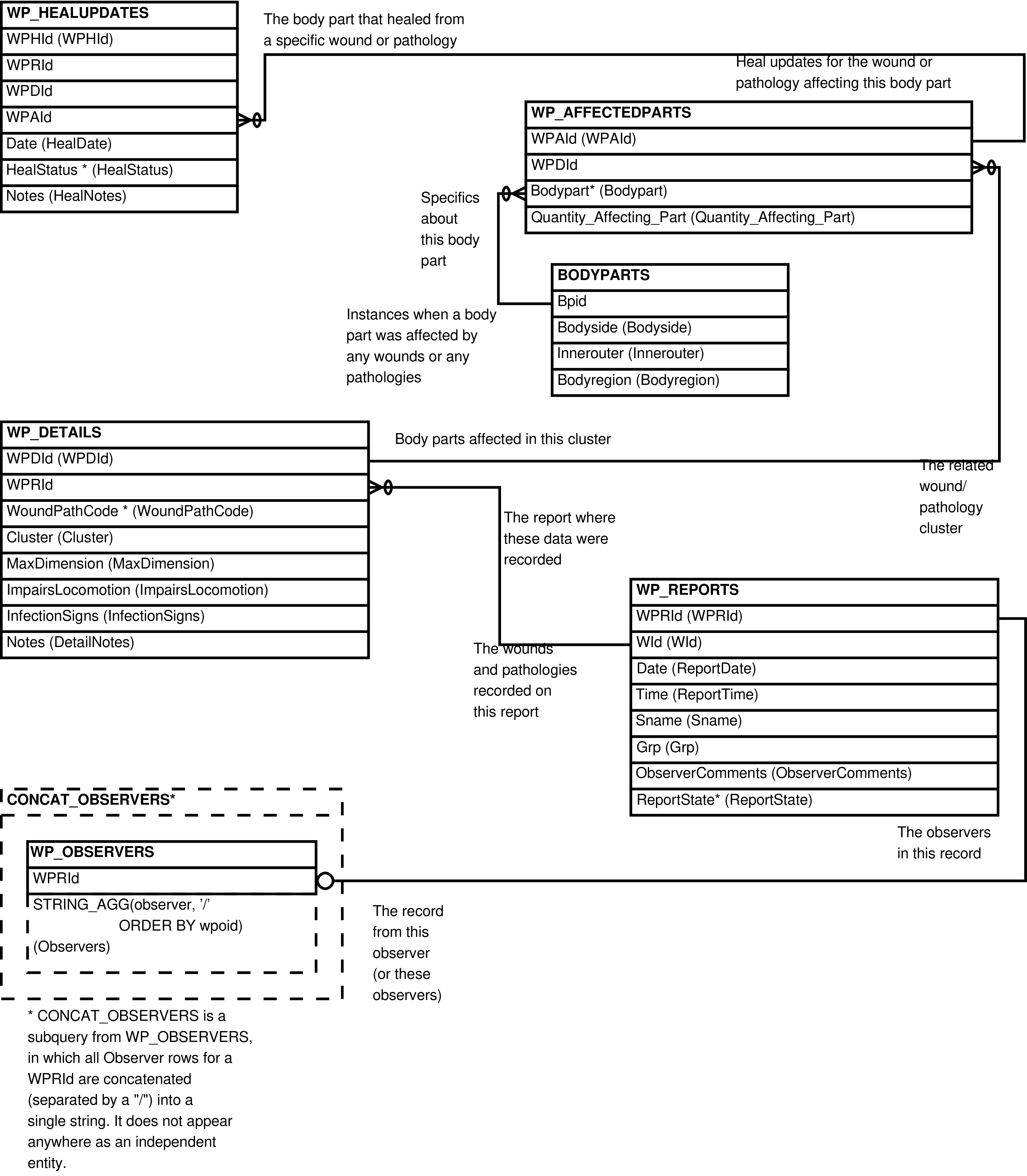 |
Table 6.22. Columns in the WP_HEALS View
| Column | From | Description |
|---|---|---|
| WPRId | WP_REPORTS.WPRId | Identifier for the report. |
| WId | WP_REPORTS.WId | User-generated identifier for the report. |
| ReportDate | WP_REPORTS.Date | The date that the report was created. |
| ReportTime | WP_REPORTS.Time | The time that the report was created. |
| Observers | WP_OBSERVERS.Observer | All of the report's observers, concatenated
together and separated by a
"/". If no
observers, then NULL. |
| Sname | WP_REPORTS.Sname | The sname of the affected individual. |
| Grp | WP_REPORTS.Grp | The group of the individual in the report, according to the observer(s). |
| ObserverComments | WP_REPORTS.ObserverComments | Notes or comments from the observer(s) about the report. |
| ReportState | WP_REPORTS.ReportState | Status of the report. |
| WPDId | WP_DETAILS.WPDId | Identifier for the wound/pathology
cluster. NULL if this heal update is only for the
report. |
| WoundPathCode | WP_DETAILS.WoundPathCode | Code indicating the wound/pathology type for
the cluster. NULL if this heal update is only for
the report. |
| Cluster | WP_DETAILS.Cluster | The wound/pathology cluster
identifier. NULL if this heal update is only for
the report. |
| MaxDimension | WP_DETAILS.MaxDimension | The highest observed length, height, depth,
etc. (as applicable), in cm, of all
wounds/pathologies in the cluster. NULL if this
heal update is only for the report. |
| ImpairsLocomotion | WP_DETAILS.ImpairsLocomotion | Boolean indicating whether or not the
wound/pathology cluster impairs the individual's
locomotion. NULL if this heal update is only for
the report. |
| InfectionSigns | WP_DETAILS.InfectionSigns | Boolean indicating whether or not the
wound/pathology cluster includes signs of an
infection. NULL if this heal update is only for
the report. |
| DetailNotes | WP_DETAILS.Notes | Textual comments or notes about the
cluster. NULL if this heal update is only for the
report. |
| WPAId | WP_AFFECTEDPARTS.WPAId | Identifier for the affected body part in the
wound/pathology cluster. NULL if this heal update
is only for the report or the cluster. |
| Bodypart | WP_AFFECTEDPARTS.Bodypart | Unique identifier for the body part. NULL
if this heal update is only for the report or the
cluster. |
| Bodyside | BODYPARTS.Bodyside | Code indicating the side of the body on which
the affected part is located. NULL if this heal
update is only for the report or the
cluster. |
| Innerouter | BODYPARTS.Innerouter | Code indicating if the affected body part is
on the inner or outer side of the body part. NULL
if this heal update is only for the report or the
cluster. |
| Bodyregion | BODYPARTS.Bodyregion | Code indicating the region on the body of the
affected body part. NULL if this heal update is
only for the report or the cluster. |
| Quantity_Affecting_Part | WP_AFFECTEDPARTS.Quantity_Affecting_Part | The number of wounds/pathologies described in
the cluster affecting this body part. NULL if this
heal update is only for the report or the
cluster. |
| WPHId | WP_HEALUPDATES.WPHId | Identifier for the heal update. |
| HealDate | WP_HEALUPDATES.Date | Date of this heal update. |
| HealStatus | WP_HEALUPDATES.HealStatus | Code indicating how well the related wound/pathology has healed. |
| HealNotes | WP_HEALUPDATES.Notes | Textual notes about the healing (or lack thereof) in this update. |
Inserting a row into WP_HEALS inserts a row into WP_HEALUPDATES, as described below.
For each row inserted into WP_HEALUPDATES, the inserted WPRId, WPDId, or WPAId value is determined based on the values provided for the other columns in this view, as described below.
To insert a WP_HEALUPDATES row
updating a report (having a
non-NULL WPRId), the
provided data must be sufficient to uniquely identify a
single row in WP_REPORTS, and should not
include any information about clusters or affected body
parts. That is, the provided values in columns from WP_REPORTS (WPRId, WId, ReportDate,
ReportTime, Sname, Grp, ObserverComments, or ReportState)
and the "Observers" must altogether be associable with a
single report, and all the columns from WP_DETAILS (WPDId, WoundPathCode, Cluster,
MaxDimension, ImpairsLocomotion, InfectionSigns, and
DetailNotes), and both WP_AFFECTEDPARTS
and BODYPARTS (WPAId, Bodypart,
Bodyside, Innerouter, Bodyregion, and
Quantity_Affecting_Part) must all be NULL. It is not
necessary to provide all of the columns from WP_REPORTS or the Observers, just enough data
to uniquely identify the report. The WPRId of the designated WP_REPORTS row is inserted as the new WP_HEALUPDATES.WPRId.
To insert a WP_HEALUPDATES row
updating a cluster (having a
non-NULL WPDId), the
provided data must be sufficient to uniquely identify a
single row in WP_DETAILS, and should not
include any information about affected body parts. That
is, the provided values in columns from WP_DETAILS (WPDId, WoundPathCode, Cluster,
MaxDimension, ImpairsLocomotion, InfectionSigns, and
DetailNotes) and WP_REPORTS (WPRId, WId,
ReportDate, ReportTime, Sname, Grp, ObserverComments, and
ReportState), and the "Observers" must altogether be
associable with a single cluster and related report, and
all the columns from both WP_AFFECTEDPARTS and BODYPARTS (WPAId, Bodypart, Bodyside,
Innerouter, Bodyregion, and Quantity_Affecting_Part) must
all be NULL. It is not necessary to provide all of the
columns from WP_DETAILS or WP_REPORTS or the Observers, just enough data
to uniquely identify the cluster. The WPDId of the designated WP_DETAILS row is inserted as the new WP_HEALUPDATES.WPDId.
To insert a WP_HEALUPDATES row
updating an affected body part
(having a non-NULL WPAId), the provided data must
be sufficient to uniquely identify a single row in WP_AFFECTEDPARTS. That is, the provided
values must altogether be associable with a single body
part, related cluster, and related report. It is not
necessary to provide all of the columns from WP_AFFECTEDPARTS, WP_DETAILS, WP_REPORTS or
the "Observers", just enough data to uniquely identify the
affected body part. The WPAId of the designated WP_AFFECTEDPARTS row is inserted as the new
WP_HEALUPDATES.WPAId.
Each new WP_HEALUPDATES row is inserted with the provided HealDate, HealStatus, and HealNotes values.
Updating a row in WP_HEALS updates the underlying columns in WP_HEALUPDATES, as described below.
Updates to the HealDate, HealStatus, and HealNotes columns update their related columns in WP_HEALUPDATES, as expected. Updates to all other columns are prohibited[250].
Tip
To update the WPRId, WPDId, or WPAId columns in a WP_HEALUPDATES row, delete the WP_HEALUPDATES row and re-enter it with updated information.
Deleting a row in WP_HEALS deletes the underlying row in WP_HEALUPDATES. Related rows in WP_REPORTS, WP_DETAILS, WP_AFFECTEDPARTS, and BODYPARTS will be unaffected.
Contains one row for every row in WP_REPORTS. In addition to all of the columns
from WP_REPORTS, this view also has an
"Observers" column showing all related WP_OBSERVERS.Observer values (if any) concatenated
together (or NULL if there are no related values).
The intended purpose of this view is for uploading data into WP_REPORTS and WP_OBSERVERS, especially multiple observers for a single report. It may also be useful for querying/accessing the data.
When
uploading data with this view, it is an error if observer
initials cannot be unambiguously interpreted. In the
admittedly-unlikely event that there is an observer whose
initials legitimately include the separator character
"/", this observer's
initials cannot be inserted via this view.[251] In this case, the offending observer code must
be removed from the data, then manually inserted into WP_OBSERVERS.
Figure 6.51. Query Defining the WP_REPORTS_OBSERVERS View
WITH concat_observers AS (SELECT wprid
, string_agg(observer, '/' ORDER BY wpoid) as observers
FROM wp_observers
GROUP BY wprid)
SELECT wp_reports.wprid AS wprid
, wp_reports.wid AS wid
, wp_reports.date AS date
, wp_reports.time AS time
, concat_observers.observers AS observers
, wp_reports.sname AS sname
, wp_reports.grp AS grp
, wp_reports.observercomments AS observercomments
, wp_reports.reportstate AS reportstate
FROM wp_reports
LEFT JOIN concat_observers
ON concat_observers.wprid = wp_reports.wprid;
Table 6.23. Columns in the WP_REPORTS_OBSERVERS View
| Column | From | Description |
|---|---|---|
| WPRId | WP_REPORTS.WPRId | Identifier for the report. |
| WId | WP_REPORTS.WId | User-generated identifier for the report. |
| Date | WP_REPORTS.Date | The date that this report was created. |
| Time | WP_REPORTS.Time | The time that this report was created. |
| Observers | WP_OBSERVERS.Observer | All of this report's related observers,
concatenated together and separated by a
"/". If no
related observers, then NULL. |
| Sname | WP_REPORTS.Sname | The sname of the affected individual. |
| Grp | WP_REPORTS.Grp | The group of the individual in this report, according to the observer(s). |
| ObserverComments | WP_REPORTS.ObserverComments | Notes or comments from the observer(s) about this report. |
| ReportState | WP_REPORTS.ReportState | Status of the report. |
Inserting a row into WP_REPORTS_OBSERVERS inserts a row into WP_REPORTS and a number of rows into WP_OBSERVERS, as described below.
Like their related columns in WP_REPORTS, the WId, Date, Sname, Grp, and
ReportState columns cannot be NULL. When there is
already a WP_REPORTS row with the
provided WPRId, WId, ReportState, Date, Time, Sname, Grp,
and ObserverComments, a new WP_REPORTS
row is not added. These values are instead used to
determine the correct WPRId to use when inserting data
into WP_OBSERVERS.
For
each "/"-separated
observer provided in the Observers column, one row is
inserted into the WP_OBSERVERS table,
with the related WPRId. A
NULL Observers column is interpreted to mean that there
are no new rows to add to WP_OBSERVERS;
it does not result in a new WP_OBSERVERS row with a NULL Observer value.
Any observer initials provided that are already present for this WPRId in WP_OBSERVERS will not be added again.
Tip
To add new observers to a report that already has some observers recorded, you can insert a row that lists all the observers--already-present and not--or a row that only lists the newly-added observers.
Updating a row in WP_REPORTS_OBSERVERS updates the underlying row in WP_REPORTS as expected, and the underlying row(s) in WP_OBSERVERS as described below.
When an update doesn't actually change the Observers column, the related data in WP_OBSERVERS are unaffected. When the update does change the Observers column, all prior rows in WP_OBSERVERS are deleted, and new rows are inserted as described above.
Deleting a row in WP_REPORTS_OBSERVERS deletes the underlying row in WP_REPORTS and the underlying rows (if any) in WP_OBSERVERS.
DELETE commands in this view
remove the WP_REPORTS row, and all
related WP_OBSERVERS rows are
deleted concomitantly. It is not possible to
only remove row(s) from WP_OBSERVERS when deleting from this
view.
Tip
To remove any observers from a report without
deleting the related WP_REPORTS row,
use the UPDATE command in this view
(see above). Alternatively, skip the view altogether
and just delete the rows directly from the WP_OBSERVERS table.
Contains one row for every row in SEXSKINS, and for every row in CYCLES that does not have a related SEXSKINS row. Each row contains the SEXSKINS columns and the related CYCLES columns. Because there is a many-to-one
relationship between SEXSKINS and CYCLES, the same CYCLES data
will appear repeatedly, once for each related SEXSKINS row. In those cases where there is CYCLES row but no related SEXSKINS row the SEXSKINS
columns will be NULL. Because a SEXSKINS
row always has a related CYCLES row, and it
is the CYCLES row that identifies the
cycling female, when working with the SEXSKINS table alone it is difficult to tell which
sexskin/PCS observations belong to which female. This view
provides a convenient way to create and maintain the SEXSKINS/CYCLES
combination.
Tip
It is usually a good idea to leave the Cid column
unspecified (NULL) when maintaining SEXSKINS using this view. This view uses the
rules described in the Sexual Cycle Determination section when the
underlying tables are maintained to automatically determine
the appropriate Cid values to use
in the SEXSKINS rows when no Cid is
supplied.
Figure 6.53. Query Defining the CYCLES_SEXSKINS View
SELECT cycles.cid AS cid
, cycles.sname AS sname
, cycles.seq AS seq
, cycles.series AS series
, sexskins.sxid AS sxid
, sexskins.date AS date
, sexskins.size AS size
, sexskins.color AS color
FROM cycles LEFT OUTER JOIN sexskins ON (cycles.cid = sexskins.cid);
Table 6.24. Columns in the CYCLES_SEXSKINS View
| Column | From | Description |
|---|---|---|
| Cid | CYCLES.Cid | Arbitrary number uniquely identifying the CYCLES row. |
| Sname | CYCLES.Sname | Female that is cycling. |
| Seq | CYCLES.Seq | Number indicating the cycle's position in time within the sequence of all the cycles of the female. Counts from 1 upwards. |
| Series | CYCLES.Series | Number indicating with which CYCLES (Female Sexual Cycles) of continuous observation the cycle belongs. |
| Sxid | SEXSKINS.Sxid | Unique number identifying the sexskin observation. |
| Date | SEXSKINS.Date | Date-of-record of the sexual cycle transition event. |
| Size | SEXSKINS.Size | Measured sexskin size. |
| Color | SEXSKINS.Color | Observed sexskin color. |
Tip
In most cases Cid, Cpid, Seq, and Series should be
unspecified (or specified as NULL), in which case Babase
will compute and assign the correct values.
Inserting a row into CYCLES_SEXSKINS or SEXSKINS_CYCLES inserts a row into SEXSKINS, as expected. A new row is never inserted into CYCLES. Either a Cid or a Sname must be supplied, it is usually preferable to supply a Sname. When a Sname is supplied Babase will determine the appropriate Cid value automatically. When a Cid is supplied and a CYCLES row already exists with the given Cid then the underlying CYCLES row is updated to conform with the inserted data.[252] Supplying a Cid serves only to identify a female. Babase automatically chooses which of a female's CYCLES to relate to the sexskin measurement based on the dates involved. For further information see the documentation of the SEXSKINS table.
Updating a row in CYCLES_SEXSKINS updates the underlying columns in CYCLES and SEXSKINS, as expected. However, the relationship between CYCLES and SEXSKINS introduces some complications.
Updating the Cid column updates[253] the Cid columns in both CYCLES and SEXSKINS.
Setting all the SEXSKINS columns
(Cid and Sxid excepted) to NULL causes the
deletion of the SEXSKINS row. Setting
SEXSKINS columns to a non-NULL value
when all the SEXSKINS columns were
NULL previously creates a new row in SEXSKINS.
Contains one row for every row in the CYCLES_SEXSKINS view. This view is sorted for ease of maintenance.
Figure 6.55. Query Defining the CYCLES_SEXSKINS_SORTED View
SELECT cycles.cid AS cid
, cycles.sname AS sname
, cycles.seq AS seq
, cycles.series AS series
, sexskins.sxid AS sxid
, sexskins.date AS date
, sexskins.size AS size
, sexskins.color AS color
FROM cycles LEFT OUTER JOIN sexskins ON (cycles.cid = sexskins.cid)
ORDER BY cycles.sname, sexskins.date;
Table 6.25. Columns in the CYCLES_SEXSKINS_SORTED View
| Column | From | Description |
|---|---|---|
| Cid | CYCLES.Cid | Arbitrary number uniquely identifying the CYCLES row. |
| Sname | CYCLES.Sname | Female that is cycling. |
| Seq | CYCLES.Seq | Number indicating the cycle's position in time within the sequence of all the cycles of the female. Counts from 1 upwards. |
| Series | CYCLES.Series | Number indicating with which CYCLES (Female Sexual Cycles) of continuous observation the cycle belongs. |
| Sxid | SEXSKINS.Sxid | Unique number identifying the sexskin observation. |
| Date | SEXSKINS.Date | Date-of-record of the sexual cycle transition event. |
| Size | SEXSKINS.Size | Measured sexskin size. |
| Color | SEXSKINS.Color | Observed sexskin color. |
The operations allowed are as described in the CYCLES_SEXSKINS view.
Contains one row for every birth or fetal loss, summarizing the reproductive event.
Caution
Pregnancies with no recorded outcome do not appear in this view. If this is a problem we can change this. (kop)
Figure 6.57. Query Defining the MATERNITIES View
SELECT cycles.sname AS mom
, cycles.cid AS cid
, cycles.seq AS seq
, cycles.series AS series
, cycpoints.cpid AS conceive
, cycpoints.date AS zdate
, members.grp AS zdate_grp
, cycpoints.edate AS edate
, cycpoints.ldate AS ldate
, cycpoints.source AS source
, pregs.pid AS pid
, pregs.parity AS parity
, biograph.bioid AS child_bioid
, biograph.sname AS child
, biograph.birth AS birth
FROM cycles
JOIN cycpoints ON (cycpoints.cid = cycles.cid)
JOIN members ON (members.date = cycpoints.date
AND members.sname = cycles.sname)
JOIN pregs ON (pregs.conceive = cycpoints.cpid)
JOIN biograph ON (pregs.pid = biograph.pid);
Table 6.26. Columns in the MATERNITIES View
| Column | From | Description |
|---|---|---|
| Mom | CYCLES.Sname | Identifier (Sname) of the mother. |
| Cid | CYCLES.Cid | Identifier of conception cycle. |
| Seq | CYCLES.Seq | Ordinal sequence of the conception cycle among all of the mother's cycles. |
| Series | CYCLES.Series | Series number. Ordinal position of the continuous period of observation during which the mother's conception cycle was recorded, among all of the periods of continuous observation of the mother. |
| Conceive | CYCPOINTS.Cpid | Identifier of the CYCPOINTS row containing the Zdate. |
| Zdate | CYCPOINTS.Date | Conception date-of-record. |
| Zdate_Grp | MEMBERS.Grp | Mother's group as of the conception date-of-record. |
| Edate | CYCPOINTS.Edate | Earliest possible date of conception. |
| Ldate | CYCPOINTS.Ldate | Latest possible date of conception. |
| Source | CYCPOINTS.Source | The origin of the conception date. This has bearing as to its accuracy. |
| Pid | PREGS.Pid | Identifier of the pregnancy. |
| Parity | PREGS.Parity | Parity of the pregnancy. |
| Child_Bioid | BIOGRAPH.Bioid | Identifier (Bioid) of the progeny. |
| Child | BIOGRAPH.Sname | Identifier (Sname) of the progeny. |
| Birth | BIOGRAPH.Birth | Birthdate of the progeny. |
Contains one row for every row in CYCLES. Each row contains the CYCLES columns and separate columns for the
related CYCPOINTS Mdate, Tdate, and Ddate
information. Sexual cycles that do not have a Mdate, Tdate,
or Ddate, where there is no such CYCPOINTS
row, contain NULL where data are missing. This view
provides a convenient way to connect the Mdates, Tdates, and
Ddates of each cycle.
Figure 6.59. Query Defining the MTD_CYCLES View
SELECT cycles.cid AS cid
, cycles.sname AS sname
, cycles.seq AS seq
, cycles.series AS series
, mcp.cpid AS mcpid
, mcp.date AS mdate
, mcp.edate AS emdate
, mcp.ldate AS lmdate
, mcp.source AS msource
, tcp.cpid AS tcpid
, tcp.date AS tdate
, tcp.edate AS etdate
, tcp.ldate AS ltdate
, tcp.source AS tsource
, dcp.cpid AS dcpid
, dcp.date AS ddate
, dcp.edate AS eddate
, dcp.ldate AS lddate
, dcp.source AS dsource
FROM cycles
LEFT OUTER JOIN cycpoints AS mcp ON (mcp.cid = cycles.cid
AND mcp.code = 'M')
LEFT OUTER JOIN cycpoints AS tcp ON (tcp.cid = cycles.cid
AND tcp.code = 'T')
LEFT OUTER JOIN cycpoints AS dcp ON (dcp.cid = cycles.cid
AND dcp.code = 'D')
ORDER BY cycles.sname, cycles.seq;
Table 6.27. Columns in the MTD_CYCLES View
| Column | From | Description |
|---|---|---|
| Cid | CYCLES.Cid | Arbitrary number uniquely identifying the CYCLES row. |
| Sname | CYCLES.Sname | Female that is cycling. |
| Seq | CYCLES.Seq | Number indicating the cycle's position in time within the sequence of all the cycles of the female. Counts from 1 upwards. |
| Series | CYCLES.Series | Number indicating with which series of continuous observation the cycle belongs. |
| Mcpid | CYCPOINTS.Cpid | Number uniquely identifying the Mdate's CYCPOINTS row, or NULL if the cycle
has no Mdate. |
| Mdate | CYCPOINTS.Date | Date-of-record of the sexual cycle's Mdate,
or NULL if the cycle has no Mdate. |
| Emdate | CYCPOINTS.Edate | Earliest possible date for the sexual cycle's
Mdate, or NULL if the cycle has no Mdate or there
is no Edate associated
with the Mdate . |
| Lmdate | CYCPOINTS.Ldate | Latest possible date for the sexual cycle's
Mdate, or NULL if the cycle has no Mdate or there
is no Ldate associated
with the Mdate. |
| Msource | CYCPOINTS.Source | Code indicating from whence the Mdate data
were derived, or NULL if the cycle has no Mdate.
This has a bearing as to its accuracy. |
| Tcpid | CYCPOINTS.Cpid | Number uniquely identifying the Tdate's CYCPOINTS row, or NULL if the cycle
has no Tdate. |
| Tdate | CYCPOINTS.Date | Date-of-record of the sexual cycle's Tdate,
or NULL if the cycle has no Tdate. |
| Etdate | CYCPOINTS.Edate | Earliest possible date for the sexual cycle's
Tdate, or NULL if the cycle has no Tdate or there
is no Edate associated
with the Tdate. |
| Ltdate | CYCPOINTS.Ldate | Latest possible date for the sexual cycle's
Tdate, or NULL if the cycle has no Tdate or there
is no Ldate associated
with the Tdate. |
| Tsource | CYCPOINTS.Source | Code indicating from whence the Tdate data
were derived, or NULL if the cycle has no Tdate.
This has a bearing as to its accuracy. |
| Dcpid | CYCPOINTS.Cpid | Number uniquely identifying the Ddate's CYCPOINTS row, or NULL if the cycle
has no Ddate. |
| Ddate | CYCPOINTS.Date | Date-of-record of the sexual cycle's Ddate,
or NULL if the cycle has no Ddate. |
| Eddate | CYCPOINTS.Edate | Earliest possible date for the sexual cycle's
Ddate, or NULL if the cycle has no Ddate or there
is no Edate associated
with the Ddate. |
| Lddate | CYCPOINTS.Ldate | Latest possible date for the sexual cycle's
Ddate, or NULL if the cycle has no Ddate or there
is no Ldate associated
with the Ddate. |
| Dsource | CYCPOINTS.Source | Code indicating from whence the Ddate data
were derived, or NULL if the cycle has no Ddate.
This has a bearing as to its accuracy. |
Inserting rows into MTD_CYCLES inserts rows into the
underlying tables as expected. However, there are
complications introduced due to the nature of the view.
No row is inserted into CYCPOINTS for a
particular Mdate, Tdate, or Ddate when the relevant Date, Edate, and Ldate columns are all NULL.
Unlike the CYCPOINTS.Source column, the
"source" columns in this view default to
D (data). Omitting a
"source" column from an INSERT statement or
specifying it as NULL results in the default value of
D.
Tip
It is strongly recommended that the Cid, Mcpid, Tcpid, and Dcpid be
assigned automatically by the system. To do this either
do not specify a value for these columns or specify a
value of NULL.
Contains one row for every row in SEXSKINS. Each row contains the SEXSKINS columns and the related CYCLES columns. Because there is a many-to-one relationship between SEXSKINS and CYCLES, the same CYCLES data will appear repeatedly, once for each related SEXSKINS row. Because a SEXSKINS row always has a related CYCLES row, and it is the CYCLES row that identifies the cycling female, when working with the SEXSKINS table alone it is difficult to tell which sexskin/PCS observations belong to which female. This view provides a convenient way to create and maintain the SEXSKINS/CYCLES combination.
Tip
It is usually a good idea to leave the Cid column
unspecified (NULL) when maintaining SEXSKINS using this view. This view uses the
rules described in the Sexual Cycle Determination section when the
underlying tables are maintained to automatically determine
the appropriate Cid values to use
in the SEXSKINS rows when no Cid is
supplied.
Note
The SEXSKINS_CYCLES view is very similar to the CYCLES_SEXSKINS view. It is unclear which is more useful so both exist.
Figure 6.61. Query Defining the SEXSKINS_CYCLES View
SELECT cycles.cid AS cid
, cycles.sname AS sname
, cycles.seq AS seq
, cycles.series AS series
, sexskins.sxid AS sxid
, sexskins.date AS date
, sexskins.size AS size
, sexskins.color AS color
FROM sexskins, cycles
WHERE cycles.cid = sexskins.cid
ORDER BY cycles.sname, sexskins.date;
Table 6.28. Columns in the SEXSKINS_CYCLES View
| Column | From | Description |
|---|---|---|
| Cid | CYCLES.Cid | Arbitrary number uniquely identifying the CYCLES row. |
| Sname | CYCLES.Sname | Female that is cycling. |
| Seq | CYCLES.Seq | Number indicating the cycle's position in time within the sequence of all the cycles of the female. Counts from 1 upwards. |
| Series | CYCLES.Series | Number indicating with which CYCLES (Female Sexual Cycles) of continuous observation the cycle belongs. |
| Sxid | SEXSKINS.Sxid | Unique number identifying the sexskin observation. |
| Date | SEXSKINS.Date | Date-of-record of the sexual cycle transition event. |
| Size | SEXSKINS.Size | Measured sexskin size. |
| Color | SEXSKINS.Color | Observed sexskin color. |
Tip
In most cases Cid, Cpid, Seq, and Series should be
unspecified (or specified as NULL), in which case Babase
will compute and assign the correct values.
Inserting a row into CYCLES_SEXSKINS or SEXSKINS_CYCLES inserts a row into SEXSKINS, as expected. A new row is never inserted into CYCLES. Either a Cid or a Sname must be supplied, it is usually preferable to supply a Sname. When a Sname is supplied Babase will determine the appropriate Cid value automatically. When a Cid is supplied and a CYCLES row already exists with the given Cid then the underlying CYCLES row is updated to conform with the inserted data.[254] Supplying a Cid serves only to identify a female. Babase automatically chooses which of a female's CYCLES to relate to the sexskin measurement based on the dates involved. For further information see the documentation of the SEXSKINS table.
Contains one row for every row in the SEXSKINS_CYCLES view. This view is sorted for ease of maintenance.
Figure 6.63. Query Defining the SEXSKINS_CYCLES_SORTED View
SELECT cycles.cid AS cid
, cycles.sname AS sname
, cycles.seq AS seq
, cycles.series AS series
, sexskins.sxid AS sxid
, sexskins.date AS date
, sexskins.size AS size
, sexskins.color AS color
FROM sexskins, cycles
WHERE cycles.cid = sexskins.cid
ORDER BY cycles.sname, sexskins.date;
Table 6.29. Columns in the SEXSKINS_CYCLES_SORTED View
| Column | From | Description |
|---|---|---|
| Cid | CYCLES.Cid | Arbitrary number uniquely identifying the CYCLES row. |
| Sname | CYCLES.Sname | Female that is cycling. |
| Seq | CYCLES.Seq | Number indicating the cycle's position in time within the sequence of all the cycles of the female. Counts from 1 upwards. |
| Series | CYCLES.Series | Number indicating with which CYCLES (Female Sexual Cycles) of continuous observation the cycle belongs. |
| Sxid | SEXSKINS.Sxid | Unique number identifying the sexskin observation. |
| Date | SEXSKINS.Date | Date-of-record of the sexual cycle transition event. |
| Size | SEXSKINS.Size | Measured sexskin size. |
| Color | SEXSKINS.Color | Observed sexskin color. |
The operations allowed are as described in the SEXSKINS_CYCLES view.
Contains one row for every date that a female has a row in SEXSKINS and/or REPRO_NOTES. Each row contains all the columns from SEXSKINS and REPRO_NOTES, and may include the Sname column from CYCLES. This view provides a convenient way to insert and maintain data from both SEXSKINS and REPRO_NOTES, as both tables' data may be entered together.
When the female has SEXSKINS data for
a date but not REPRO_NOTES, the columns
exclusive to REPRO_NOTES — RNId and
Note — will be NULL. When she has REPRO_NOTES data but not SEXSKINS, the columns exclusive to SEXSKINS — Cid, Sxid, Size, and Color
— will be NULL.
The source of the Sname and Date columns depends on whether the female has data in SEXSKINS for the row's Date. If yes, the Sname is the related CYCLES.Sname and the Date is the SEXSKINS.Date. If no — and she does have data in REPRO_NOTES for this date[256] — the Sname and Date are the REPRO_NOTES.Sname and Date.
Tip
It is usually a good idea to leave the Cid column
unspecified (NULL) when maintaining SEXSKINS using this view. This view uses the
rules described in the Sexual Cycle Determination section when the
underlying tables are maintained to automatically determine
the appropriate Cid values to use
in the SEXSKINS rows when no Cid is
supplied.
Figure 6.65. Query Defining the SEXSKINS_REPRO_NOTES View
SELECT COALESCE(cycles.sname, repro_notes.sname) AS sname
, COALESCE(sexskins.date, repro_notes.date) AS date
, sexskins.cid AS cid
, sexskins.sxid AS sxid
, sexskins.size AS size
, sexskins.color AS color
, repro_notes.rnid AS rnid
, repro_notes.note AS note
FROM sexskins
JOIN cycles
ON cycles.cid = sexskins.cid
FULL OUTER JOIN repro_notes
ON repro_notes.sname = cycles.sname
AND repro_notes.date = sexskins.date;
Table 6.30. Columns in the SEXSKINS_REPRO_NOTES View
| Column | From | Description |
|---|---|---|
| Sname |
| Female under observation. |
| Date |
| Date of observation. |
| Cid | SEXSKINS.Cid | Unique identifier for the related CYCLES row, or NULL if there are no
rows in SEXSKINS for this
Date. |
| Sxid | SEXSKINS.Sxid | Unique identifier for the sexskin
observation, or NULL if there are no rows in SEXSKINS for this Date. |
| Size | SEXSKINS.Size | Observed sexskin size, or NULL if there are
no rows in SEXSKINS for this
Date. |
| Color | SEXSKINS.Color | Observed sexskin color, or NULL if there
are no rows in SEXSKINS for this
Date. |
| RNId | REPRO_NOTES.RNId | Unique identifier for the reproductive note,
or NULL if there are no rows in REPRO_NOTES for this Date. |
| Note | REPRO_NOTES.Note | Text of the reproductive note, or NULL if
there are no rows in REPRO_NOTES
for this Date. |
Tip
In most cases the Cid should be unspecified (or
specified as NULL), in which case Babase will compute
and assign the correct value.
Inserting a row into SEXSKINS_REPRO_NOTES inserts rows into SEXSKINS and/or REPRO_NOTES, as expected. Either a Cid or Sname must be supplied, but it is usually preferable to supply the Sname. When Sname is supplied, Babase will determine the appropriate Cid value automatically.
When all of the columns exclusive to SEXSKINS — Cid, Sxid, Size, and Color
— are NULL, the view will not attempt to insert a
row into SEXSKINS.
When both of the columns exclusive to REPRO_NOTES — RNId and Note — are
NULL, the view will not attempt to insert a row into
REPRO_NOTES.
Each insert to this view must insert something
somewhere. It is an error for all of
the table-exclusive columns listed above to be
NULL.
Updating a row in SEXSKINS_REPRO_NOTES updates the underlying columns in SEXSKINS and REPRO_NOTES (if any), as expected.
Deleting a row from SEXSKINS_REPRO_NOTES deletes the underlying columns from SEXSKINS and REPRO_NOTES (if any), as expected.
Contains one row for every row in INTERACT_DATA. Each row contains a column for the
actor and a column for the actee. The actor and actee are
retrieved from the PARTS table, when there
is no related parts row the actor or actee is NULL.
This view is somewhat useful for the maintenance and analysis of social interaction data.[257] It's primarily optimized for speed and so finds its best use when writing queries.
This view indicates whether
each interaction can be considered "representative".
Representative interactions are those collected as part of a
"representative interactions" protocol, which involves
observers collecting all interactions in their line of sight
in the course of collecting focal samples on all individuals
of a given age-sex class in a random order. The observer
cannot possibly record all the
interactions that occur in the group, but the interactions
collected this way are considered a representative
sample of all the interactions. For each
interaction, if there are any focal samples (in SAMPLES) that i) are on the same Date, ii) are
with the same Observer, iii) are in the same group (Actor_Grp
or Actee_Grp), iv) have at least one point in-sight and v) follow a
representative interactions protocol, then that interaction
can be considered representative and its Repr_Interxn will be
TRUE. Only representative interactions can be used to
calculate the relative frequencies with which interactions
occur.
Note
It is still possible to see all-occurrences data for focal individuals, as described earlier.
Tip
Resist the urge to think of the "representative" label as an indication of the quality of the observation. If the quality of an observation were in question, it would not be added to Babase at all.
Figure 6.67. Query Defining the ACTOR_ACTEES View
WITH sampling AS (SELECT samples.date
, samples.observer
, ARRAY_AGG(DISTINCT members.grp) AS grps
FROM samples
JOIN stypes
ON stypes.stype = samples.stype
AND stypes.repr_interxns
JOIN members
ON members.sname = samples.sname
AND members.date = samples.date
WHERE samples.minsis > 0
GROUP BY samples.date, samples.observer)
SELECT interact_data.iid AS iid
, interact_data.sid AS sid
, interact_data.act AS act
, interact_data.date AS date
, interact_data.start AS start
, interact_data.stop AS stop
, interact_data.observer AS observer
, actor.partid AS actorid
, COALESCE(actor.sname, '998'::CHAR(3)) AS actor
, actorms.grp AS actor_grp
, actee.partid AS acteeid
, COALESCE(actee.sname, '998'::CHAR(3)) AS actee
, acteems.grp AS actee_grp
, interact_data.handwritten AS handwritten
, interact_data.exact_date AS exact_date
, COALESCE((actorms.grp = ANY(sampling.grps)
OR acteems.grp = ANY(sampling.grps))
, FALSE) AS repr_interxn
FROM interact_data
LEFT OUTER JOIN parts AS actor
ON (actor.iid = interact_data.iid AND actor.role = 'R')
LEFT OUTER JOIN parts AS actee
ON (actee.iid = interact_data.iid AND actee.role = 'E')
LEFT OUTER JOIN members AS actorms
ON (actorms.sname = actor.sname
AND actorms.date = interact_data.date)
LEFT OUTER JOIN members AS acteems
ON (acteems.sname = actee.sname
AND acteems.date = interact_data.date)
LEFT OUTER JOIN sampling
ON (sampling.date = interact_data.date
AND sampling.observer = interact_data.observer);
Table 6.31. Columns in the ACTOR_ACTEES View
| Column | From | Description |
|---|---|---|
| Iid | INTERACT_DATA.Iid | Identifier of the interaction. |
| Sid | INTERACT_DATA.Sid | Identifier of the sample, if any, during which the data was collected. |
| Act | INTERACT_DATA.Act | The kind of interaction. |
| Date | INTERACT_DATA.Date | The date of the interaction. |
| Start | INTERACT_DATA.Start | The time the interaction began. |
| Stop | INTERACT_DATA.Stop | The time the interaction ended. |
| Observer | INTERACT_DATA.Observer | The observer who recorded the interaction. |
| Actorid | PARTS.Partid | The Partid of the actor's PARTS row. |
| Actor | PARTS.Sname | The Sname of the
actor in the interaction, when there is a PARTS row for the actor. Otherwise, the
value 998. |
| Actor_Grp | MEMBERS.Grp | The Grp of the actor on the date of the interaction. |
| Acteeid | PARTS.Partid | The Partid of the actee's PARTS row. |
| Actee | PARTS.Sname | The Sname of the
actee in the interaction, when there is a PARTS row for the actee. Otherwise, the
value 998. |
| Actee_Grp | MEMBERS.Grp | The Grp of the actee on the date of the interaction. |
| Handwritten | INTERACT_DATA.Handwritten | Whether or not the interaction was recorded in handwritten records. |
| Exact_Date | INTERACT_DATA.Exact_Date | Whether this row's Date is the specific day
that the interaction occurred (TRUE) or only the
year and month of the interaction (FALSE). |
| Repr_Interxn | Subquery — see Figure 6.68. | Whether this Observer performed any randomized focal samples in this group (Actor_Grp or Actee_Grp) on this Date |
The Actor_Grp and Actee_Grp columns are computed.
Attempts to put a value into these columns raise an error if
the new value is not NULL or does not correspond to the
computed value.
The Repr_Interxns column is also computed. Attempts to put a value in this column are silently ignored.
Tip
Best practice is to omit computed columns from inserts and updates.
The Actor and Actee columns must not be NULL or
998 when inserting
into or updating this view.
When inserting or updating ACTOR_ACTEES the values of
the Actor_Grp and Actee_Grp columns must either be NULL or
match the group recorded for the individual for that day in
MEMBERS, if such a row exists. Inserting
and updating ACTOR_ACTEES cannot affect group
membership.
Tip
It is usually a good idea to leave the computed
columns unspecified (NULL) when maintaining social
interactions using this view.
Inserting a row into ACTOR_ACTEES inserts a row into INTERACT_DATA and two rows, one for the actor and one for the actee, into PARTS, as expected.
To insert an actor without an actee (or vice versa)
use a NULL value for the Actee (or Actor).
Tip
When entering a new social interaction it is
usually a good idea to leave Iid unspecified (or
specified as NULL). In this case Babase will compute
a new Iid and use it
appropriately in the new PARTS rows.
Likewise the Actorid and Acteeid columns are usually
best left NULL, in which case Babase will also create
appropriate values.
Updating a row in ACTOR_ACTEES updates the underlying columns in INTERACT_DATA and PARTS, as expected.
An actor or actees PARTS row can
be deleted or inserted by UPDATE of ACTOR_ACTEES. To
insert an new PARTS row supply a Sname for the actor or
actee where there was none. To delete a actor or or actee
set the Sname to NULL.
When deleting an actor or actee either the
corresponding Actorid/Acteeid value must be set to NULL,
or the corresponding Actorid/Acteeid value must be
unaltered.
Tip
The Actor and Actee cannot be switched with a update operation.[258] Delete the interaction and re-create it instead.
Deleting a row in ACTOR_ACTEES deletes the underlying row in INTERACT_DATA and the two underlying rows in PARTS, as expected.
Contains one row for every row in INTERACT_DATA. There is no difference between this view and the INTERACT_DATA table, other than the view extends the INTERACT_DATA table with additional date and time columns that transform the corresponding INTERACT_DATA columns in useful and interesting ways, and the view is sorted by Iid.
Figure 6.69. Query Defining the INTERACT View
SELECT iid AS iid
, interact_data.sid AS sid
, interact_data.act AS act
, acts.class AS class
, interact_data.date AS date
, julian(interact_data.date) AS jdate
, interact_data.start AS start
, spm(interact_data.start) AS startspm
, stop AS stop
, spm(interact_data.stop) AS stopspm
, interact_data.observer AS observer
, interact_data.handwritten AS handwritten
, interact_data.exact_date AS exact_date
FROM interact_data
JOIN acts
ON (acts.act = interact_data.act);
Table 6.32. Columns in the INTERACT View
| Column | From | Description |
|---|---|---|
| Iid | INTERACT_DATA.Iid | Identifier of the interaction. |
| Sid | INTERACT_DATA.Sid | Identifier of the point observation collection, if any, during which the data was collected. |
| Act | INTERACT_DATA.Act | The kind of interaction. |
| Date | INTERACT_DATA.Date | The date of the interaction. |
| Jdate | INTERACT_DATA.Date (computed) | The date of the interaction, in Julian date form. |
| Start | INTERACT_DATA.Start | The time the interaction began. |
| Startspm | INTERACT_DATA.Start (computed) | The time the interaction begin (Start), represented as the number of seconds past midnight. This is useful for computing and analyzing time intervals outside of the PostgreSQL environment. |
| Stop | INTERACT_DATA.Stop | The time the interaction ended. |
| Stopspm | INTERACT_DATA.Stop (computed) | The time the interaction ended (Stop), represented as the number of seconds past midnight. This is useful for computing and analyzing time intervals outside of the PostgreSQL environment. |
| Observer | INTERACT_DATA.Observer | The observer who recorded the interaction. |
| Handwritten | INTERACT_DATA.Handwritten | Whether or not the interaction was recorded in handwritten records. |
| Exact_Date | INTERACT_DATA.Exact_Date | Whether this row's Date is the specific day
that the interaction occurred
(TRUE) or only the year and month
of the interaction (FALSE). |
Warning
Any modifications to the Jdate, Startspm, or Stopspm columns are silently ignored.
Inserting a row into INTERACT inserts a row into INTERACT_DATA, as expected.
Updating a row in INTERACT updates the underlying columns in INTERACT_DATA, as expected.
Deleting a row in INTERACT deletes the underlying row in INTERACT_DATA.
Contains one row for every row in the INTERACT view. There is no difference between this view and the INTERACT view, other than that this view is sorted by Iid.
Figure 6.71. Query Defining the INTERACT_SORTED View
SELECT iid AS iid
, interact_data.sid AS sid
, interact_data.act AS act
, acts.class AS class
, interact_data.date AS date
, julian(interact_data.date) AS jdate
, interact_data.start AS start
, spm(interact_data.start) AS startspm
, interact_data.stop AS stop
, spm(interact_data.stop) AS stopspm
, interact_data.observer AS observer
, interact_data.handwritten AS handwritten
, interact_data.exact_date AS exact_date
FROM interact_data
JOIN acts
ON (acts.act = interact_data.act)
ORDER BY iid;
Table 6.33. Columns in the INTERACT_SORTED View
| Column | From | Description |
|---|---|---|
| Iid | INTERACT_DATA.Iid | Identifier of the interaction. |
| Sid | INTERACT_DATA.Sid | Identifier of the point observation collection, if any, during which the data was collected. |
| Act | INTERACT_DATA.Act | The kind of interaction. |
| Date | INTERACT_DATA.Date | The date of the interaction. |
| Jdate | INTERACT_DATA.Date (computed) | The date of the interaction, in Julian date form. |
| Start | INTERACT_DATA.Start | The time the interaction began. |
| Startspm | INTERACT_DATA.Start (computed) | The time the interaction begin (Start), represented as the number of seconds past midnight. This is useful for computing and analyzing time intervals outside of the PostgreSQL environment. |
| Stop | INTERACT_DATA.Stop | The time the interaction ended. |
| Stopspm | INTERACT_DATA.Stop (computed) | The time the interaction ended (Stop), represented as the number of seconds past midnight. This is useful for computing and analyzing time intervals outside of the PostgreSQL environment. |
| Observer | INTERACT_DATA.Observer | The observer who recorded the interaction. |
| Handwritten | INTERACT_DATA.Handwritten | Whether or not the interaction was recorded in handwritten records. |
| Exact_Date | INTERACT_DATA.Exact_Date | Whether this row's Date is the specific day
that the interaction occurred
(TRUE) or only the year and month
of the interaction (FALSE). |
The operations allowed are as described in the INTERACT view.
Contains one row for every row in MPI_DATA. Each row contains a column for the
actor and a column for the actee. The actor and actee are
retrieved from the MPI_PARTS table, when
there is no related parts row the actor or actee is NULL.
The MPIS table supplies the date of the
interaction and there are further self-joins to the MPI_DATA, MPIACTS, and MPI_PARTS tables to compute help request
circumstances and outcome.
This view is useful for the analysis of multiparty interaction data.[259].
Figure 6.73. Query Defining the MPI_EVENTS View
SELECT mpis.mpiid AS mpiid
, mpis.date AS date
, mpis.observer AS observer
, mpis.context_type AS context_type
, mpis.context AS context
, mpi_data.mpidid AS mpidid
, mpi_data.seq AS seq
, mpi_data.mpiact AS mpiact
, actor.mpipid AS actorid
, actor.sname AS actor
, actor.unksname AS unkactor
, actee.mpipid AS acteeid
, actee.sname AS actee
, actee.unksname AS unkactee
, CASE WHEN EXISTS(SELECT 1
FROM mpiacts
WHERE mpiacts.mpiact = mpi_data.mpiact
AND mpiacts.kind = 'H')
THEN
EXISTS(SELECT 1
FROM mpi_data AS request
, mpiacts
, mpi_parts AS requestor
, mpi_parts AS requestee
WHERE request.mpiid = mpi_data.mpiid
AND request.seq < mpi_data.seq
AND mpiacts.mpiact = request.mpiact
AND mpiacts.kind = 'R'
AND requestor.mpidid = request.mpidid
AND requestor.role = 'R'
AND requestor.sname = actee.sname
AND requestee.mpidid = request.mpidid
AND requestee.role = 'E'
AND requestee.sname = actor.sname)
ELSE
NULL
END AS solicited
, EXISTS(SELECT 1
FROM mpi_data AS initial,
mpiacts
WHERE initial.mpiid = mpi_data.mpiid
AND initial.seq = 1
AND mpiacts.mpiact = initial.mpiact
AND mpiacts.decided)
AS decided
, mpi_data.helped AS helped
, mpi_data.active AS active
FROM mpis
LEFT OUTER JOIN mpi_data ON (mpis.mpiid = mpi_data.mpiid)
LEFT OUTER JOIN mpi_parts AS actor
ON (actor.mpidid = mpi_data.mpidid AND actor.role = 'R')
LEFT OUTER JOIN mpi_parts AS actee
ON (actee.mpidid = mpi_data.mpidid AND actee.role = 'E');
Table 6.34. Columns in the MPI_EVENTS View
| Column | From | Description |
|---|---|---|
| Mpiid | MPIS.Mpiid | Identifier of the multiparty interaction collection. |
| Date | MPIS.Date | The date of the multiparty interaction collection. |
| Observer | MPIS.Observer | The initials of the observer of the multiparty interaction. |
| Context_type | MPIS.Context_type | The context type of the multiparty interaction collection. |
| Context | MPIS.MPIS-Context | Text describing the context of the multiparty interaction collection. |
| Mpidid | MPI_DATA.Mpidid | Identifier of the dyadic interaction. |
| Seq | MPI_DATA.Seq | Number that orders the dyadic interaction in
time within the multiparty interaction collection.
The first dyadic interaction has a Seq value of
1, the second a Seq value of
2, etc. Identical Seq values
indicate interactions which occurred
simutainously. |
| Mpiact | MPI_DATA.MPIAct | The kind of dyadic interaction. |
| Actorid | MPI_PARTS.Mpipid | The Mpipid of the actor's MPI_PARTS row. |
| Actor | MPI_PARTS.Sname | The Sname of the actor in the interaction. |
| Unkactor | MPI_PARTS.Unksname | The PARTUNKS.Unksname code which denotes why the actor is unknown. |
| Acteeid | MPI_PARTS.Mpipid | The Mpipid of the actee's MPI_PARTS row. |
| Actee | MPI_PARTS.Sname | The Sname of the actee in the interaction. |
| Unkactee | MPI_PARTS.Unksname | The PARTUNKS.Unksname code which denotes why the actee is unknown. |
| Solicited | Computed | A boolean: TRUE or FALSE, or NULL.
NULL when the act is not an act of help (MPIACTS.MPIAct
is FALSE). Whether or not the help given was
solicited with a request for help -- whether MPIACTS.Kind
is R, from a
previous (having a smaller MPI_DATA.Seq)
MPI act's MPI_DATA.MPIAct value where the actor
and actee match. For more details see the Figure 6.73. |
| Decided | Computed | A boolean: TRUE or FALSE. Whether or not
the result of the MPI was decided when the event took
place -- obtained from the MPIACTS.Decided value of the MPI_DATA.MPIAct of the first (MPI_DATA.Seq =
1) interaction of the MPI. |
| Helped | MPI_DATA.Helped | Whether or not help was given in response to
the request for help. NULL when the event is not a
request for help. |
| Active | MPI_DATA.Active | Whether the help given was active or passive.
NULL when the event is not a request for help or no
help was forthcoming. |
Caution
Attempts to change the computed columns, Solicited and Decided, are silently ignored.
Tip
It is usually a good idea to leave the computed
columns, as well as the automatically assigned ID columns,
unspecified (NULL) when maintaining social interactions
using this view.
Inserting a row into MPI_EVENTs inserts a row into MPI_DATA and two rows, one for the actor and one for the actee, into MPI_PARTS, as expected. It also may insert a row into the MPIS table.
Warning
The presence or absence of a Mpiid value
determines whether or not a MPIS row
is created. When Mpiid is NULL (or the column is not
specified) a new MPIS row is created.
When a non-NULL value is supplied the given Mpiid
identifies the existing multiparty interaction
collection, a MPIS row, to which the
new dyadic interaction is added.
Warning
The value of the Date, Context_type, and MPIS-Context columns are ignored when the supplied Mpiid identifies an existing MPIS row.
Warning
The PostgreSQL nextval()
function cannot be part of an INSERT
expression which assigns a value to this view's Mpiid or
Mpidid columns.
Tip
When entering a new social interaction it is
usually a good idea to leave Mpidid unspecified (or
specified as NULL). In this case Babase will compute
a new Mpiid and use it
appropriately in the new MPI_PARTS
rows. Likewise the Actorid and Acteeid columns are
usually best left NULL, in which case Babase will also
create appropriate values.
Updating a row in MPI_EVENTS updates the underlying columns in MPIS, MPI_DATA, and MPI_PARTS as expected.
Tip
Because updates to the database occur one table at a time, and because Babase does not allow an interaction to have the same individual as both the actor and actee, it is impossible to switch the Actor and Actee with a update operation. Delete the interaction and re-create it instead.
This view returns no rows, it is used only to upload
multiparty interaction data into the MPIS,
MPI_DATA, MPI_PARTS, and
CONSORTS tables. Attempting to
SELECT rows from this view will raise an
error.
This view exists instead of a custom upload program.
Each line in the uploaded file corresponds to one or more dyadic interactions. Each multiparty interaction is represented in the input file by contiguous lines, with these lines ordered so that earlier interactions appear first in the file. The context of the multiparty interaction and the result of any consort context must appear on the first line, and only the first line, of those uploaded lines that make up the MPI.
A single line in the file usually corresponds to a
single dyadic interaction. The exception is when the first
line of the multiparty interaction has an MPI_DATA.MPIAct value
indicating that multiple initial interactions are allowed.
In this case the row represents multiple dyadic
interactions, one for each combination of the Snames in the actor
and actee columns. For example, if there
are 3 actors and 2 actees there will be a total of
6[260] dyadic interactions.
The uploaded file may contain leading or trailing empty lines. No data must be indicated by an empty cell.
The uploaded file must begin with a line of column headings with the names given below in the order given below. The column headings are validated but otherwise unused. This is to assist in the detection of data entry errors. The content of each column is as described.
midA number that identifies the row within the uploaded file. These numbers must increase with each row and must be sequential within any one multiparty interaction. Gaps are allowed between multiparty interactions.
This column must contain a value.
This data is not recorded in the database but is checked for validity to assist in detection of data entry errors.
coal_idA number that identifies the coalition within the uploaded file. All the rows associated with a given multiparty interaction must share the same number. These numbers must not otherwise be re-used within the uploaded file.[261]
This column must contain a value.
This data is not recorded in the database but is checked for validity to assist in detection of data entry errors.
grpA GROUPS.Gid value. This data is not recorded in the database but is checked to ensure that each input line for a given multiparty interaction contains the same value. This check is done to assist in detection of data entry errors. The data in this column is not otherwise validated.
dateThe date of the multiparty interaction. This data is stored in the MPIS.Date database column.
All the rows associated with a given multiparty interaction must contain the same date value. This check is done to assist in detection of data entry errors.
observerThe initials of the observer of the multiparty interaction. This data is stored in the MPIS.Observer database column.
All the rows associated with a given multiparty interaction must contain the same observer value. This check is done to assist in detection of data entry errors.
actorThe Snames of the actor(s) that is/are interacting. When there is more than one actor (see above) the Snames of the actors are separated by a comma (
,).This data is stored in the MPI_PARTS.Sname database column, unless the value is is one of those in PARTUNKS.Unksname in which case it is stored in the MPI_PARTS.Unksname column.
agg_actA code indicating the act performed. These codes are generally MPIACTS values, with the following exceptions.[262]
+A
+is changed intoAH.?A
?is changed intoRE.PA
Pis changed into aPH.
recipThe Snames of the actee(s) that is/are interacting. When there is more than one actee (see above) the Snames of the actees are separated by a comma (
,).This data is stored in the MPI_PARTS.Sname database column, unless the value is is one of those in PARTUNKS.Unksname in which case it is stored in the MPI_PARTS.Unksname column.
outcomeThe result of a request for help. The allowed values are:
- (blank)
Indicates no data -- the action was not a request for help. A blank entry results in
NULLvalues for MPI_DATA.Helped and MPI_DATA.Active.SUCCIndicates that active help was given in response to the help request. MPI_DATA.Helped and MPI_DATA.Active are set to
TRUE.FAILIndicates an unsuccessful request for help. MPI_DATA.Helped and MPI_DATA.Active are set to
FALSE.PASSIndicates that passive help was given in response to the help request. MPI_DATA.Helped is set to
TRUEand MPI_DATA.Active set toFALSE.
form_passive_aidThe values in this column are ignored.
contextThe MPIS.MPIS-Context value. A value may only appear on the first line of the lines making up the multiparty interaction. When the context_type is
Cthe context value must beCONSORTand aNULLwill be the value entered into the database. This check is done to assist detection of data entry errors.consortA record of the result of consortship context, if any. A value may only appear on the first line of the lines making up the multiparty interaction. If not blank the consort value has the form: “male1 WITH female;male2 GET female”, or the form “male1 WITH female;male2 KEEP female”. In either case mpi_upload checks to see that both occurrences in the “female” placeholder are identical. When any of the participants are unknown the individual should be a PARTUNKS.Unksname value. When the “KEEP” form is used mpi_upload checks to see that the “male1” and “male2” values are identical.[263]
The “male1” value is recorded in the CONSORTS.Had database column. The “male2” value is recorded in the CONSORTS.Got database column.
context_typeThe MPIS.Context_type code. A value may only appear on the first line of the lines making up the multiparty interaction.
Figure 6.75. Query Defining the MPI_UPLOAD View
SELECT NULL::INT AS mid
, NULL::INT AS coal_id
, NULL::TEXT AS grp
, NULL::date AS date
, NULL::TEXT AS observer
, NULL::TEXT AS actor
, NULL::TEXT AS agg_act
, NULL::TEXT AS recip
, NULL::TEXT AS outcome
, NULL::TEXT AS form_passive_aid
, NULL::TEXT AS context
, NULL::TEXT AS consort
, NULL::TEXT AS context_type
WHERE _raise_babase_exception(
'Cannot select MPI_UPLOAD'
|| ': The only use of the MPI_UPLOAD view is to insert'
|| ' new data into the MPI portion of babase');
Figure 6.76. Entity Relationship Diagram of the MPI_UPLOAD View
Contains one row for every row in POINT_DATA. There is no difference between this view and the POINT_DATA table, other than the view contains additional columns that may be useful derivatives of the Ptime column.
Tip
Use this view instead of the POINT_DATA table.
Figure 6.77. Query Defining the POINTS View
SELECT pntid AS pntid
, sid AS sid
, activity AS activity
, posture AS posture
, foodcode AS foodcode
, ptime AS ptime
, spm(ptime) AS ptimespm
FROM point_data;
Table 6.35. Columns in the POINTS View
| Column | From | Description |
|---|---|---|
| Pntid | POINT_DATA.Pntid | Identifier of the point observation. |
| Sid | POINT_DATA.Sid | Identifier of the sample during which the data was collected. |
| Activity | POINT_DATA.Activity | The kind of activity the focal was engaged in. |
| Posture | POINT_DATA.Posture | The posture of the focal. |
| Foodcode | POINT_DATA.Foodcode | The food eaten, if any. |
| Ptime | POINT_DATA.Ptime | The time the observation was recorded, with a precision of 1 second. |
| Ptimespm | POINT_DATA.Ptime (computed) | The time the point observation was recorded (Ptime) represented as the number of seconds past midnight. This is useful for computing and analyzing time intervals outside of the PostgreSQL environment. |
The Ptimespm column is computed. Attempts to put a
value into the Ptimespm column raise an error if the new
value is not NULL or does not correspond to the computed
value.
Tip
Best practice is to omit computed columns from inserts and updates.
Inserting a row into POINTS inserts a row into POINT_DATA, as expected.
Updating a row in POINTS updates the underlying columns in POINT_DATA, as expected.
Deleting a row in POINTS deletes the underlying row in POINT_DATA.
Contains one row for every row in POINTS. There is no difference between this view and the POINTS view, other than this view is sorted by Sid, and within that by Ptime.
Figure 6.79. Query Defining the POINTS_SORTED View
SELECT pntid AS pntid
, sid AS sid
, activity AS activity
, posture AS posture
, foodcode AS foodcode
, ptime AS ptime
, ptimespm AS ptimespm
FROM points
ORDER BY sid, ptime;
Table 6.36. Columns in the POINTS_SORTED View
| Column | From | Description |
|---|---|---|
| Pntid | POINT_DATA.Pntid | Identifier of the point observation. |
| Sid | POINT_DATA.Sid | Identifier of the sample during which the data was collected. |
| Activity | POINT_DATA.Activity | The kind of activity the focal was engaged in. |
| Posture | POINT_DATA.Posture | The posture of the focal. |
| Foodcode | POINT_DATA.Foodcode | The food eaten, if any. |
| Ptime | POINT_DATA.Ptime | The time the observation was recorded, with a precision of 1 second. |
| Ptimespm | POINT_DATA.Ptime (computed) | The time the point observation was recorded (Ptime) represented as the number of seconds past midnight. This is useful for computing and analyzing time intervals outside of the PostgreSQL environment. |
The operations allowed are as described in the POINTS view.
Contains one row for every row in SAMPLES. This row is identical to the SAMPLES table except that it has an additional column Grp_of_focal which contains the group of the focal on the sampling date.[264]
Figure 6.81. Query Defining the SAMPLES_GOFF View
SELECT samples.sid AS sid
, samples.date AS date
, samples.stime AS stime
, samples.observer AS observer
, samples.stype AS stype
, samples.grp AS grp
, samples.sname AS sname
, samples.mins AS mins
, samples.minsis AS minsis
, samples.programid AS programid
, samples.setupid AS setupid
, samples.collection_system AS collection_system
, members.grp AS grp_of_focal
FROM members, samples
WHERE members.sname = samples.sname
AND members.date = CAST(samples.date AS DATE);
Table 6.37. Columns in the SAMPLES_GOFF View
| Column | From | Description |
|---|---|---|
| Sid | SAMPLES.Sid | Identifier of the sample. |
| Date | SAMPLES.Date | Date of sample collection. |
| STime | SAMPLES.Stime | Time of sample collection. |
| Observer | SAMPLES.Observer | Observer who collected the sample. |
| SType | SAMPLES.SType | A code indicating the nature of the focal individual and the data collection procedure used. |
| Grp | SAMPLES.Grp | The group the observation team sampled. |
| Sname | SAMPLES.Sname | Identifier of sampled individual. |
| Mins | SAMPLES.Mins | Sample duration in minutes, from start to finish. |
| Minsis | SAMPLES.Minsis | Number of minutes of sample data. |
| Programid | SAMPLES.Programid | Identifer of the software ("program", if any) used with this row's Collection_System to collect the focal sample. |
| Setupid | SAMPLES.Setupid | The configuration file (if any) used by this row's Programid to collect this sample's data. |
| Collection_System | SAMPLES.Collection_System | The device or hardware configuration used to collect the sample. |
| Grp_of_focal | MEMBERS.Grp | The group of the sampled individual. |
Contains one row for every unique Dartid value in the ANESTHS table.[265] Each row statistically summarizes the ANESTHS rows having the common Dartid value.
This view is useful when joined with the DARTINGS table on Dartid.
Figure 6.83. Query Defining the ANESTH_STATS View
SELECT anesths.dartid AS dartid
, count(*) AS ansamps
, avg(anesths.anamount) AS anamount_mean
, stddev(anesths.anamount) AS anamount_stddev
FROM anesths
GROUP BY anesths.dartid;
Table 6.38. Columns in the ANESTH_STATS View
| Column | From | Description |
|---|---|---|
| Dartid | ANESTHS.Dartid | Identifier of the darting event. |
| Ansamps | Computed | Number of ANESTHS rows having the given Dartid value -- the number of times additional anesthetic was administered during the darting. |
| Anamount_mean | ANESTHS.Anamount (computed) | The arithmetic mean of the additional anesthetic amounts related to the given Dartid -- the mean of the additional anesthetic administered during the darting. |
| Anamount_stddev | ANESTHS.Anamount (computed) | The standard deviation of the additional anesthetic amounts related to the given Dartid -- the standard deviation of the additional anesthetic administered during the darting. |
Contains one row for every unique Dartid value in the BODYTEMPS table.[266] Each row statistically summarizes the BODYTEMPS rows having the common Dartid value.
This view is useful when joined with the DARTINGS table on Dartid.
Figure 6.85. Query Defining the BODYTEMP_STATS View
SELECT bodytemps.dartid AS dartid
, count(*) AS btsamps
, avg(bodytemps.btemp) AS btemp_mean
, stddev(bodytemps.btemp) AS btemp_stddev
FROM bodytemps
GROUP BY bodytemps.dartid;
Table 6.39. Columns in the BODYTEMP_STATS View
| Column | From | Description |
|---|---|---|
| Dartid | BODYTEMPS.Dartid | Identifier of the darting event. |
| Btsamps | Computed | Number of BODYTEMPS rows having the given Dartid value -- the number of body temperature measurements taken during the darting. |
| Btemp_mean | BODYTEMPS.Btemp (computed) | The arithmetic mean of the body temperature measurements related to the given Dartid -- the mean of the body temperature measurements taken during the darting. |
| Btemp_stddev | BODYTEMPS.Btemp (computed) | The standard deviation of the body temperature measurements related to the given Dartid -- the standard deviation of the body temperature measurements taken during the darting. |
Contains one row for every unique Dartid value in the CHESTS table.[267] Each row statistically summarizes the CHESTS rows having the common Dartid value.
This view is useful when joined with the DARTINGS table on Dartid.
Figure 6.87. Query Defining the CHEST_STATS View
SELECT chests.dartid AS dartid
, count(*) AS chsamps
, avg(chests.chcircum) AS chcircum_mean
, stddev(chests.chcircum) AS chcircum_stddev
, avg(chests.chunadjusted) AS chunadjusted_mean
, stddev(chests.chunadjusted) AS chunadjusted_stddev
FROM chests
GROUP BY chests.dartid;
Table 6.40. Columns in the CHEST_STATS View
| Column | From | Description |
|---|---|---|
| Dartid | CHESTS.Dartid | Identifier of the darting event. |
| Chsamps | Computed | Number of CHESTS rows having the given Dartid value -- the number of chest circumference measurements taken during the darting. |
| Chcircum_mean | CHESTS.Chcircum (computed) | The arithmetic mean of the chest circumference measurements related to the given Dartid -- the mean of the chest circumference measurements taken during the darting. |
| Chcircum_stddev | CHESTS.Chcircum (computed) | The standard deviation of the chest circumference measurements related to the given Dartid -- the standard deviation of the chest circumference measurements taken during the darting. |
| Chunadjusted_mean | CHESTS.Chunadjusted (computed) | The arithmetic mean of the unadjusted chest circumference measurements related to the given Dartid -- the mean of the unadjusted chest circumference measurements taken during the darting. |
| Chunadjusted_stddev | CHESTS.Chunadjusted (computed) | The standard deviation of the unadjusted chest circumference measurements related to the given Dartid -- the standard deviation of the unadjusted chest circumference measurements taken during the darting. |
Contains one row for every unique Dartid value in the CROWNRUMPS table.[268] Each row statistically summarizes the CROWNRUMPS rows having the common Dartid value.
This view is useful when joined with the DARTINGS table on Dartid.
Figure 6.89. Query Defining the CROWNRUMP_STATS View
SELECT crownrumps.dartid AS dartid
, count(*) AS crsamps
, avg(crownrumps.crlength) AS crlength_mean
, stddev(crownrumps.crlength) AS crlength_stddev
FROM crownrumps
GROUP BY crownrumps.dartid;
Table 6.41. Columns in the CROWNRUMP_STATS View
| Column | From | Description |
|---|---|---|
| Dartid | CROWNRUMPS.Dartid | Identifier of the darting event. |
| CRsamps | Computed | Number of CROWNRUMPS rows having the given Dartid value -- the number of crown-to-rump measurements taken during the darting. |
| CRlength_mean | CROWNRUMPS.CRlength (computed) | The arithmetic mean of the crown-to-rump measurements related to the given Dartid -- the mean of the crown-to-rump measurements taken during the darting. |
| CRlength_stddev | CROWNRUMPS.CRlength (computed) | The standard deviation of the crown-to-rump measurements related to the given Dartid -- the standard deviation of the crown-to-rump measurements taken during the darting. |
This view is used to insert data about the proportional presence of specific cell types in a blood sample as determined by flow cytometric analysis.
Each row inserted into this view represents an analysis of a single blood sample. That is, a single row in the FLOW_CYTOMETRY table.
The Name, Sname, Sex, and Dartdate columns are not inserted anywhere. Instead, they are used to identify the darting — that is, to identify the appopriate DARTINGS.Dartid — to which all the row's data belong. As discussed below, it is important that the darting event be added to DARTINGS before adding data via this view.
Figure 6.91. Query Defining the DART_FLOW_CYTOMETRY_UPLOAD View
SELECT NULL::TEXT AS name
, NULL::TEXT AS sname
, NULL::TEXT AS sex
, NULL::TEXT AS dartdate
, NULL::TEXT AS flow_date
, NULL::TEXT AS monocytes
, NULL::TEXT AS nk
, NULL::TEXT AS b
, NULL::TEXT AS helper_t
, NULL::TEXT AS cytotoxic_t
, NULL::TEXT AS comments
WHERE _raise_babase_exception(
'Cannot select DART_FLOW_CYTOMETRY_UPLOAD'
|| ': The only use of the DART_FLOW_CYTOMETRY_UPLOAD view is to insert'
|| ' new data into the FLOW_CYTOMETRY table');
Figure 6.92. Entity Relationship Diagram of the DART_FLOW_CYTOMETRY_UPLOAD View
Table 6.42. Columns in the DART_FLOW_CYTOMETRY_UPLOAD View
| Column | Uploads into | Description |
|---|---|---|
| Name | Nowhere; used to determine this darting's Dartid | The BIOGRAPH.Name associated with this darting's Sname. |
| Sname | Nowhere; used to determine this darting's Dartid | This darting's Sname. |
| Sex | Nowhere; used to determine this darting's Dartid | The BIOGRAPH.Sex associated with this darting's Sname. |
| Dartdate | Nowhere; used to determine this darting's Dartid | The Date of the darting. |
| Flow_Date | FLOW_CYTOMETRY.Flow_Date | The date of the flow cytometric analysis. |
| Monocytes | FLOW_CYTOMETRY.Monocytes | The percentage of PBMCs in this sample that were identified as monocytes. |
| NK | FLOW_CYTOMETRY.NK | The percentage of PBMCs in this sample that were identified as natural killer cells. |
| B | FLOW_CYTOMETRY.B | The percentage of PBMCs in this sample that were identified as B cells. |
| Helper_T | FLOW_CYTOMETRY.Helper_T | The percentage of PBMCs in this sample that were identified as helper T cells. |
| Cytotoxic_T | FLOW_CYTOMETRY.Cytotoxic_T | The percentage of PBMCs in this sample that were identified as cytotoxic T cells. |
| Comments | FLOW_CYTOMETRY.Comments | Comments or miscellaneous information about this analysis. |
Only INSERT is allowed on DART_FLOW_CYTOMETRY_UPLOAD; SELECT, UPDATE, and DELETE are not allowed. Inserting a row into DART_FLOW_CYTOMETRY_UPLOAD inserts a row into FLOW_CYTOMETRY as described above.
This view is used to insert data about the logistics of a darting, e.g. who was darted, when, which drug was used and how much, was additional anesthetic applied after the initial dart, etc.
Each row inserted into this view represents a single darting. The values in each of the columns are used to insert a new row into DARTINGS, and into one or more new rows in ANESTHS.
This view includes three sets of three "extra_anesthN" columns, e.g. extra_anesth1, extra_anesth_time1, and extra_anesth_amt1. Each of these sets of three is used to describe a single administration of "extra" anasthetic after the initial dart, and is inserted as a new row in ANESTHS. Thus, a single row inserted in this view may include the addition of up to 3 rows in that table.
When one column of an "extra_anesthN" set is non-NULL,
the others must also be non-NULL.
Caution
For any given darting, the logistic data must be uploaded first. After that, data may be inserted into the other DART_xxx_UPLOAD views in any order.
Figure 6.93. Query Defining the DART_LOGISTICS_UPLOAD View
SELECT NULL::TEXT AS name
, NULL::TEXT AS sname
, NULL::TEXT AS sex
, NULL::TEXT AS dartdate
, NULL::TEXT AS darttime
, NULL::TEXT AS downtime
, NULL::TEXT AS pickuptime
, NULL::TEXT AS dartdrug
, NULL::TEXT AS extra_anesth1
, NULL::TEXT AS extra_anesth_time1
, NULL::TEXT AS extra_anesth_amt1
, NULL::TEXT AS extra_anesth2
, NULL::TEXT AS extra_anesth_time2
, NULL::TEXT AS extra_anesth_amt2
, NULL::TEXT AS extra_anesth3
, NULL::TEXT AS extra_anesth_time3
, NULL::TEXT AS extra_anesth_amt3
, NULL::TEXT AS other_notes
, NULL::TEXT AS comments
WHERE _raise_babase_exception(
'Cannot select DART_LOGISTICS_UPLOAD'
|| ': The only use of the DART_LOGISTICS_UPLOAD view is to insert'
|| ' new data into its related tables');
Figure 6.94. Entity Relationship Diagram of the DART_LOGISTICS_UPLOAD View
Table 6.43. Columns in the DART_LOGISTICS_UPLOAD View
| Column | Uploads into | Description |
|---|---|---|
| Name | Nowhere; used to validate the provided Sname value | The BIOGRAPH.Name associated with the provided Sname. It is an error if not. |
| Sname | DARTINGS.Sname | The BIOGRAPH.Sname of the darted individual. |
| Sex | Nowhere; used to validate the provided Sname value | The BIOGRAPH.Sex associated with the provided Sname. It is an error if not. |
| Dartdate | DARTINGS.Date | The date the individual was darted. |
| Darttime | DARTINGS.Darttime | The time the individual was darted. |
| Downtime | DARTINGS.Downtime | The time the individual succumbed to the anesthetic. |
| Pickuptime | DARTINGS.Pickuptime | The time the individual was picked up by the darting team. |
| Dartdrug | DARTINGS.Drug | The DRUGS.Drug of the anesthetic delivered by the dart. |
| Extra_Anesth1 | ANESTHS.Drug | If extra anesthetic was administered, the DRUGS.Drug of that anesthetic. |
| Extra_Anesth_Time1 | ANESTHS.Antime | If extra anasthetic was administered, the time of the administration. |
| Extra_Anesth_Amt1 | ANESTHS.Anamount | If extra anasthetic was administered, the amount of anesthetic administered. |
| Extra_Anesth2 | ANESTHS.Drug | If a second round of extra anesthetic was administered, the DRUGS.Drug of that anesthetic. |
| Extra_Anesth_Time2 | ANESTHS.Antime | If a second round of extra anasthetic was administered, the time of the administration. |
| Extra_Anesth_Amt2 | ANESTHS.Anamount | If a second round of extra anasthetic was administered, the amount of anesthetic administered. |
| Extra_Anesth3 | ANESTHS.Drug | If a third round of extra anesthetic was administered, the DRUGS.Drug of that anesthetic. |
| Extra_Anesth_Time3 | ANESTHS.Antime | If a third round of extra anasthetic was administered, the time of the administration. |
| Extra_Anesth_Amt3 | ANESTHS.Anamount | If a third round of extra anasthetic was administered, the amount of anesthetic administered. |
| Other_Notes | DARTINGS.Logisticnotes | Textual notes related to darting logistics. |
| Comments | DARTINGS.Dartcomments | General comments on the darting. |
This view is used to insert data about morphology of a darted individual, e.g. body mass, chest circumference, ulna length, etc.
Each row inserted into this view represents a single darting. The values in each of the columns may be used to update data in DARTINGS and insert new rows into CROWNRUMPS, CHESTS, ULNAS, and HUMERUSES.
This view includes several sets of columns that allow the addition of multiple rows to the same table, e.g. Crownrump1 and Crobserver1, Crownrump2 and Crobserver2, etc. The column sets related to CROWNRUMPS are two columns each, while the sets for the other tables are three columns each. Each of these sets is used to describe a single measurement, and is inserted as a new row in its related table. Thus, a single row inserted in this view may include the addition of many rows in each related table.
The Name, Sname, Sex, and Dartdate columns are not inserted anywhere. Instead, they are used to identify the darting — that is, to identify the appopriate DARTINGS.Dartid — to which all the row's data belong. As discussed earlier, it is important that the darting event be added to DARTINGS before adding data via this view.
Figure 6.95. Query Defining the DART_MORPHOLOGY_UPLOAD View
SELECT NULL::TEXT AS name
, NULL::TEXT AS sname
, NULL::TEXT AS sex
, NULL::TEXT AS dartdate
, NULL::TEXT AS bodymass
, NULL::TEXT AS crownrump1
, NULL::TEXT AS crobserver1
, NULL::TEXT AS crownrump2
, NULL::TEXT AS crobserver2
, NULL::TEXT AS crownrump3
, NULL::TEXT AS crobserver3
, NULL::TEXT AS crownrump4
, NULL::TEXT AS crobserver4
, NULL::TEXT AS crownrump5
, NULL::TEXT AS crobserver5
, NULL::TEXT AS crownrump6
, NULL::TEXT AS crobserver6
, NULL::TEXT AS chestcircum1
, NULL::TEXT AS unadj_chestcircum1
, NULL::TEXT AS chobserver1
, NULL::TEXT AS chestcircum2
, NULL::TEXT AS unadj_chestcircum2
, NULL::TEXT AS chobserver2
, NULL::TEXT AS chestcircum3
, NULL::TEXT AS unadj_chestcircum3
, NULL::TEXT AS chobserver3
, NULL::TEXT AS chestcircum4
, NULL::TEXT AS unadj_chestcircum4
, NULL::TEXT AS chobserver4
, NULL::TEXT AS chestcircum5
, NULL::TEXT AS unadj_chestcircum5
, NULL::TEXT AS chobserver5
, NULL::TEXT AS chestcircum6
, NULL::TEXT AS unadj_chestcircum6
, NULL::TEXT AS chobserver6
, NULL::TEXT AS ulna1
, NULL::TEXT AS unadj_ulna1
, NULL::TEXT AS ulobserver1
, NULL::TEXT AS ulna2
, NULL::TEXT AS unadj_ulna2
, NULL::TEXT AS ulobserver2
, NULL::TEXT AS ulna3
, NULL::TEXT AS unadj_ulna3
, NULL::TEXT AS ulobserver3
, NULL::TEXT AS ulna4
, NULL::TEXT AS unadj_ulna4
, NULL::TEXT AS ulobserver4
, NULL::TEXT AS ulna5
, NULL::TEXT AS unadj_ulna5
, NULL::TEXT AS ulobserver5
, NULL::TEXT AS ulna6
, NULL::TEXT AS unadj_ulna6
, NULL::TEXT AS ulobserver6
, NULL::TEXT AS humerus1
, NULL::TEXT AS unadj_humerus1
, NULL::TEXT AS huobserver1
, NULL::TEXT AS humerus2
, NULL::TEXT AS unadj_humerus2
, NULL::TEXT AS huobserver2
, NULL::TEXT AS humerus3
, NULL::TEXT AS unadj_humerus3
, NULL::TEXT AS huobserver3
, NULL::TEXT AS humerus4
, NULL::TEXT AS unadj_humerus4
, NULL::TEXT AS huobserver4
, NULL::TEXT AS humerus5
, NULL::TEXT AS unadj_humerus5
, NULL::TEXT AS huobserver5
, NULL::TEXT AS humerus6
, NULL::TEXT AS unadj_humerus6
, NULL::TEXT AS huobserver6
, NULL::TEXT AS crnotes
, NULL::TEXT AS chnotes
, NULL::TEXT AS ulnotes
, NULL::TEXT AS hunotes
WHERE _raise_babase_exception(
'Cannot select DART_MORPHOLOGY_UPLOAD'
|| ': The only use of the DART_MORPHOLOGY_UPLOAD view is to insert'
|| ' new data into its related tables');
Figure 6.96. Entity Relationship Diagram of the DART_MORPHOLOGY_UPLOAD View
Table 6.44. Columns in the DART_MORPHOLOGY_UPLOAD View
| Column | Uploads into | Description |
|---|---|---|
| Name | Nowhere; used to determine this darting's Dartid | The BIOGRAPH.Name associated with this darting's Sname. |
| Sname | Nowhere; used to determine this darting's Dartid | This darting's Sname. |
| Sex | Nowhere; used to determine this darting's Dartid | The BIOGRAPH.Sex associated with this darting's Sname. |
| Dartdate | Nowhere; used to determine this darting's Dartid | The Date of the darting. |
| Bodymass | DARTINGS.Mass | The individual's mass, if it was recorded. |
| Crownrump1 | CROWNRUMPS.CRlength | The first crown-to-rump measurement, if it was recorded. |
| Crobserver1 | CROWNRUMPS.CRobserver | The observer who took the first crownrump measurement, if known. |
| Crownrump2 | CROWNRUMPS.CRlength | The second crown-to-rump measurement, if it was recorded. |
| Crobserver2 | CROWNRUMPS.CRobserver | The observer who took the second crownrump measurement, if known. |
| Crownrump3 | CROWNRUMPS.CRlength | The third crown-to-rump measurement, if it was recorded. |
| Crobserver3 | CROWNRUMPS.CRobserver | The observer who took the third crownrump measurement, if known. |
| Crownrump4 | CROWNRUMPS.CRlength | The fourth crown-to-rump measurement, if it was recorded. |
| Crobserver4 | CROWNRUMPS.CRobserver | The observer who took the fourth crownrump measurement, if known. |
| Crownrump5 | CROWNRUMPS.CRlength | The fifth crown-to-rump measurement, if it was recorded. |
| Crobserver5 | CROWNRUMPS.CRobserver | The observer who took the fifth crownrump measurement, if known. |
| Crownrump6 | CROWNRUMPS.CRlength | The sixth crown-to-rump measurement, if it was recorded. |
| Crobserver6 | CROWNRUMPS.CRobserver | The observer who took the sixth crownrump measurement, if known. |
| Chestcircum1 | CHESTS.Chcircum | The first chest circumference measurement, if it was recorded. |
| Unadj_Chestcircum1 | CHESTS.Chunadjusted | The first chest circumference measurement, unadjusted, if any. |
| Chobserver1 | CHESTS.Chobserver | The observer of the first chest circumference measurement, if known. |
| Chestcircum2 | CHESTS.Chcircum | The second chest circumference measurement, if it was recorded. |
| Unadj_Chestcircum2 | CHESTS.Chunadjusted | The second chest circumference measurement, unadjusted, if any. |
| Chobserver2 | CHESTS.Chobserver | The observer of the second chest circumference measurement, if known. |
| Chestcircum3 | CHESTS.Chcircum | The third chest circumference measurement, if it was recorded. |
| Unadj_Chestcircum3 | CHESTS.Chunadjusted | The third chest circumference measurement, unadjusted, if any. |
| Chobserver3 | CHESTS.Chobserver | The observer of the third chest circumference measurement, if known. |
| Chestcircum4 | CHESTS.Chcircum | The fourth chest circumference measurement, if it was recorded. |
| Unadj_Chestcircum4 | CHESTS.Chunadjusted | The fourth chest circumference measurement, unadjusted, if any. |
| Chobserver4 | CHESTS.Chobserver | The observer of the fourth chest circumference measurement, if known. |
| Chestcircum5 | CHESTS.Chcircum | The fifth chest circumference measurement, if it was recorded. |
| Unadj_Chestcircum5 | CHESTS.Chunadjusted | The fifth chest circumference measurement, unadjusted, if any. |
| Chobserver5 | CHESTS.Chobserver | The observer of the fifth chest circumference measurement, if known. |
| Chestcircum6 | CHESTS.Chcircum | The sixth chest circumference measurement, if it was recorded. |
| Unadj_Chestcircum6 | CHESTS.Chunadjusted | The sixth chest circumference measurement, unadjusted, if any. |
| Chobserver6 | CHESTS.Chobserver | The observer of the sixth chest circumference measurement, if known. |
| Ulna1 | ULNAS.Ullength | The first ulna length measurement, if it was recorded. |
| Unadj_Ulna1 | ULNAS.Ulunadjusted | The first ulna length measurement, unadjusted, if any. |
| Ulobserver1 | ULNAS.Ulobserver | The observer of the first ulna length measurement, if it was recorded. |
| Ulna2 | ULNAS.Ullength | The second ulna length measurement, if it was recorded. |
| Unadj_Ulna2 | ULNAS.Ulunadjusted | The second ulna length measurement, unadjusted, if any. |
| Ulobserver2 | ULNAS.Ulobserver | The observer of the second ulna length measurement, if it was recorded. |
| Ulna3 | ULNAS.Ullength | The third ulna length measurement, if it was recorded. |
| Unadj_Ulna3 | ULNAS.Ulunadjusted | The third ulna length measurement, unadjusted, if any. |
| Ulobserver3 | ULNAS.Ulobserver | The observer of the third ulna length measurement, if it was recorded. |
| Ulna4 | ULNAS.Ullength | The fourth ulna length measurement, if it was recorded. |
| Unadj_Ulna4 | ULNAS.Ulunadjusted | The fourth ulna length measurement, unadjusted, if any. |
| Ulobserver4 | ULNAS.Ulobserver | The observer of the fourth ulna length measurement, if it was recorded. |
| Ulna5 | ULNAS.Ullength | The fifth ulna length measurement, if it was recorded. |
| Unadj_Ulna5 | ULNAS.Ulunadjusted | The fifth ulna length measurement, unadjusted, if any. |
| Ulobserver5 | ULNAS.Ulobserver | The observer of the fifth ulna length measurement, if it was recorded. |
| Ulna6 | ULNAS.Ullength | The sixth ulna length measurement, if it was recorded. |
| Unadj_Ulna6 | ULNAS.Ulunadjusted | The sixth ulna length measurement, unadjusted, if any. |
| Ulobserver6 | ULNAS.Ulobserver | The observer of the sixth ulna length measurement, if it was recorded. |
| Humerus1 | HUMERUSES.Hulength | The first humerus length measurement, if it was recorded. |
| Unadj_Humerus1 | HUMERUSES.Huunadjusted | The first humerus length measurement, unadjusted, if any. |
| Huobserver1 | HUMERUSES.Huobserver | The observer of the first humerus length measurement, if it was recorded. |
| Humerus2 | HUMERUSES.Hulength | The second humerus length measurement, if it was recorded. |
| Unadj_Humerus2 | HUMERUSES.Huunadjusted | The second humerus length measurement, unadjusted, if any. |
| Huobserver2 | HUMERUSES.Huobserver | The observer of the second humerus length measurement, if it was recorded. |
| Humerus3 | HUMERUSES.Hulength | The third humerus length measurement, if it was recorded. |
| Unadj_Humerus3 | HUMERUSES.Huunadjusted | The third humerus length measurement, unadjusted, if any. |
| Huobserver3 | HUMERUSES.Huobserver | The observer of the third humerus length measurement, if it was recorded. |
| Humerus4 | HUMERUSES.Hulength | The fourth humerus length measurement, if it was recorded. |
| Unadj_Humerus4 | HUMERUSES.Huunadjusted | The fourth humerus length measurement, unadjusted, if any. |
| Huobserver4 | HUMERUSES.Huobserver | The observer of the fourth humerus length measurement, if it was recorded. |
| Humerus5 | HUMERUSES.Hulength | The fifth humerus length measurement, if it was recorded. |
| Unadj_Humerus5 | HUMERUSES.Huunadjusted | The fifth humerus length measurement, unadjusted, if any. |
| Huobserver5 | HUMERUSES.Huobserver | The observer of the fifth humerus length measurement, if it was recorded. |
| Humerus6 | HUMERUSES.Hulength | The sixth humerus length measurement, if it was recorded. |
| Unadj_Humerus6 | HUMERUSES.Huunadjusted | The sixth humerus length measurement, unadjusted, if any. |
| Huobserver6 | HUMERUSES.Huobserver | The observer of the sixth humerus length measurement, if it was recorded. |
| CRnotes | DARTINGS.CRnotes | Notes on the crown-to-rump measurements. |
| Chnotes | DARTINGS.Chnotes | Notes on the chest circumference measurements. |
| Ulnotes | DARTINGS.Ulnotes | Notes on the ulna length measurements. |
| Hunotes | DARTINGS.Hunotes | Notes on the humerus length measurements. |
Only INSERT is allowed on DART_MORPHOLOGY_UPLOAD; SELECT, UPDATE, and DELETE are not allowed. Inserting a row into DART_MORPHOLOGY_UPLOAD updates DARTINGS and inserts rows into multiple tables as described above.
This view is used to insert data about the physiology of a darted individual, e.g. packed cell volume, body temperature, pulse, etc.
Each row inserted into this view represents a single darting. The values in each of the columns may be used to update data in DARTINGS, insert a new row into DPHYS, and insert multiple new rows into PCVS and BODYTEMPS.
This view includes some columns or sets of columns that allow the addition of multiple rows to the same table. Specifically, each of the hematocritN columns is inserted in its own row in PCVS; and each bodytempN-bodytemptimeN pair is inserted in its own row in BODYTEMPS. Thus, a single row inserted in this view may include the addition of many rows in each related table.
The Name, Sname, Sex, and Dartdate columns are not inserted anywhere. Instead, they are used to identify the darting — that is, to identify the appopriate DARTINGS.Dartid — to which all the row's data belong. As discussed earlier, it is important that the darting event be added to DARTINGS before adding data via this view.
Figure 6.97. Query Defining the DART_PHYSIOLOGY_UPLOAD View
SELECT NULL::TEXT AS name
, NULL::TEXT AS sname
, NULL::TEXT AS sex
, NULL::TEXT AS dartdate
, NULL::TEXT AS hematocrit1
, NULL::TEXT AS hematocrit2
, NULL::TEXT AS hematocrit3
, NULL::TEXT AS bodytemp1
, NULL::TEXT AS bodytemptime1
, NULL::TEXT AS bodytemp2
, NULL::TEXT AS bodytemptime2
, NULL::TEXT AS bodytemp3
, NULL::TEXT AS bodytemptime3
, NULL::TEXT AS pulse
, NULL::TEXT AS respiration
, NULL::TEXT AS r_inguinal_lymph
, NULL::TEXT AS l_inguinal_lymph
, NULL::TEXT AS r_axillary_lymph
, NULL::TEXT AS l_axillary_lymph
, NULL::TEXT AS r_submandibular_lymph
, NULL::TEXT AS l_submandibular_lymph
, NULL::TEXT AS other_notes_measures
, NULL::TEXT AS pcvnotes
, NULL::TEXT AS bodytempnotes
WHERE _raise_babase_exception(
'Cannot select DART_PHYSIOLOGY_UPLOAD'
|| ': The only use of the DART_PHYSIOLOGY_UPLOAD view is to insert'
|| ' new data into its related tables');
Figure 6.98. Entity Relationship Diagram of the DART_PHYSIOLOGY_UPLOAD View
Table 6.45. Columns in the DART_PHYSIOLOGY_UPLOAD View
| Column | Uploads into | Description |
|---|---|---|
| Name | Nowhere; used to determine this darting's Dartid | The BIOGRAPH.Name associated with this darting's Sname. |
| Sname | Nowhere; used to determine this darting's Dartid | This darting's Sname. |
| Sex | Nowhere; used to determine this darting's Dartid | The BIOGRAPH.Sex associated with this darting's Sname. |
| Dartdate | Nowhere; used to determine this darting's Dartid | The Date of the darting. |
| Hematocrit1 | PCVS.PCV | The first measurement of the individual's packed cell volume, if it was recorded. |
| Hematocrit2 | PCVS.PCV | The second measurement of the individual's packed cell volume, if it was recorded. |
| Hematocrit3 | PCVS.PCV | The third measurement of the individual's packed cell volume, if it was recorded. |
| Bodytemp1 | BODYTEMPS.Btemp | The first measurement of the individual's body temperature, if it was recorded. |
| Bodytemptime1 | BODYTEMPS.Bttime | The time that the first body temperature measurement was collected, if it was recorded. |
| Bodytemp2 | BODYTEMPS.Btemp | The second measurement of the individual's body temperature, if it was recorded. |
| Bodytemptime2 | BODYTEMPS.Bttime | The time that the second body temperature measurement was collected, if it was recorded. |
| Bodytemp3 | BODYTEMPS.Btemp | The third measurement of the individual's body temperature, if it was recorded. |
| Bodytemptime3 | BODYTEMPS.Bttime | The time that the third body temperature measurement was collected, if it was recorded. |
| Pulse | DPHYS.Pulse | The individual's pulse, if it was recorded. |
| Respiration | DPHYS.Respiration | The individual's respiration, if it was recorded. |
| R_Inguinal_Lymph | DPHYS.Ringnode | State of the individual's right inguinal lymph node, if it was recorded. |
| L_Inguinal_Lymph | DPHYS.Lingnode | State of the individual's left inguinal lymph node, if it was recorded. |
| R_Axillary_Lymph | DPHYS.Raxnode | State of the individual's right axillary lymph node, if it was recorded. |
| L_Axillary_Lymph | DPHYS.Laxnode | State of the individual's left axillary lymph node, if it was recorded. |
| R_Submandibular_Lymph | DPHYS.Rsubmandnode | State of the individual's right submandibular lymph node, if it was recorded. |
| L_Submandibular_Lymph | DPHYS.Lsubmandnode | State of the individual's left submandibular lymph node, if it was recorded. |
| Other_Notes_Measures | DARTINGS.Dphysnotes | Notes on physiological features, if any. |
| PCVnotes | DARTINGS.PCVnotes | Notes on the PCV measurements, if any. |
| Bodytempnotes | DARTINGS.Bodytempnotes | Notes on the body temperature measurements, if any. |
This view is used to record the tissue samples collected during a darting, including both the sample type and the quantity collected.
Each row inserted into this view represents a specific sample type collected during a specific darting, and is inserted as single row into DART_SAMPLES. The related DARTINGS.Dsamplenotes for the specified darting may also be updated.
The Name, Sname, Sex, and Dartdate columns are not inserted anywhere. Instead, they are used to identify the darting — that is, to identify the appopriate DARTINGS.Dartid — to which all the row's data belong. As discussed earlier, it is important that the darting event be added to DARTINGS before adding data via this view.
This view will not overwrite data in DARTINGS.Dsamplenotes.
If a row has a non-NULL Dsamplenotes, an error will be
returned if the darting's Dsamplenotes is already non-NULL. This
may help prevent the addition of duplicate data.
Figure 6.99. Query Defining the DART_SAMPLES_UPLOAD View
SELECT NULL::TEXT AS name
, NULL::TEXT AS sname
, NULL::TEXT AS sex
, NULL::TEXT AS dartdate
, NULL::TEXT AS type_descr
, NULL::TEXT AS num
, NULL::TEXT AS dsamplenotes
WHERE _raise_babase_exception(
'Cannot select DART_SAMPLES_UPLOAD'
|| ': The only use of the DART_SAMPLES_UPLOAD view is to insert'
|| ' new data into its related tables');
Figure 6.100. Entity Relationship Diagram of the DART_SAMPLES_UPLOAD View
Table 6.46. Columns in the DART_SAMPLES_UPLOAD View
| Column | Uploads into | Description |
|---|---|---|
| Name | Nowhere; used to determine this darting's Dartid | The BIOGRAPH.Name associated with this darting's Sname. |
| Sname | Nowhere; used to determine this darting's Dartid | This darting's Sname. |
| Sex | Nowhere; used to determine this darting's Dartid | The BIOGRAPH.Sex associated with this darting's Sname. |
| Dartdate | Nowhere; used to determine this darting's Dartid | The Date of the darting. |
| Type_Descr | Nowhere; used to determine this row's DART_SAMPLES.DS_Type | The DART_SAMPLE_TYPES.Descr of this row's sample type |
| Num | DART_SAMPLES.Num | The number of samples of this type that were collected. |
| Dsamplenotes | DARTINGS.Dsamplenotes | Notes or comments about the samples or sample collection. |
Only INSERT is allowed on DART_SAMPLES_UPLOAD; SELECT, UPDATE, and DELETE are not allowed. Inserting a row into DART_SAMPLES_UPLOAD inserts a row into the DART_SAMPLES table as described above. It may also update a row in the DARTINGS table, as described below.
When the inserted row has a non-NULL Dsamplenotes,
the related DARTINGS.Dsamplenotes for this darting is
updated with that value.
Caution
When multple rows from the same darting each have different Dsamplenotes values, each row's Dsamplenotes value replaces the notes from the previous row. Thus, if a single darting has comments/notes from multiple sample types, those comments should all be collated into the Dsamplenotes of a single row before insertion.
This view is used to insert data about the presence/absence and condition of a darted individual's teeth.
Each row inserted into this view represents a single darting. The values in each of the columns may be used to update data in DARTINGS and to insert data into TEETH. A single row inserted into this view may result in the addition of many rows into TEETH.
The Name, Sname, Sex, and Dartdate columns are not inserted anywhere. Instead, they are used to identify the darting — that is, to identify the appopriate DARTINGS.Dartid — to which all the row's data belong. As discussed earlier, it is important that the darting event be added to DARTINGS before adding data via this view.
This view includes a pair of columns for each tooth
recorded in TOOTHCODES: one for the tooth's
"state" and another for the tooth's "condition". The name of
each of these columns begins with its related Toothcode value. For example, given that
the Toothcode for the first left
upper incisor is lui1, the two
columns containing data for that tooth are
lui1_tstate and lui1_tcondition, and
those columns' values can be used to insert a new TEETH row with Tooth =
lui1.
Figure 6.101. Query Defining the DART_TEETH_UPLOAD View
SELECT NULL::TEXT AS name
, NULL::TEXT AS sname
, NULL::TEXT AS sex
, NULL::TEXT AS dartdate
, NULL::TEXT AS rum3_tstate
, NULL::TEXT AS rum3_tcondition
, NULL::TEXT AS rum2_tstate
, NULL::TEXT AS rum2_tcondition
, NULL::TEXT AS rum1_tstate
, NULL::TEXT AS rum1_tcondition
, NULL::TEXT AS rup2_tstate
, NULL::TEXT AS rup2_tcondition
, NULL::TEXT AS rup1_tstate
, NULL::TEXT AS rup1_tcondition
, NULL::TEXT AS ruc_tstate
, NULL::TEXT AS ruc_tcondition
, NULL::TEXT AS rui2_tstate
, NULL::TEXT AS rui2_tcondition
, NULL::TEXT AS rui1_tstate
, NULL::TEXT AS rui1_tcondition
, NULL::TEXT AS lui1_tstate
, NULL::TEXT AS lui1_tcondition
, NULL::TEXT AS lui2_tstate
, NULL::TEXT AS lui2_tcondition
, NULL::TEXT AS luc_tstate
, NULL::TEXT AS luc_tcondition
, NULL::TEXT AS lup1_tstate
, NULL::TEXT AS lup1_tcondition
, NULL::TEXT AS lup2_tstate
, NULL::TEXT AS lup2_tcondition
, NULL::TEXT AS lum1_tstate
, NULL::TEXT AS lum1_tcondition
, NULL::TEXT AS lum2_tstate
, NULL::TEXT AS lum2_tcondition
, NULL::TEXT AS lum3_tstate
, NULL::TEXT AS lum3_tcondition
, NULL::TEXT AS llm3_tstate
, NULL::TEXT AS llm3_tcondition
, NULL::TEXT AS llm2_tstate
, NULL::TEXT AS llm2_tcondition
, NULL::TEXT AS llm1_tstate
, NULL::TEXT AS llm1_tcondition
, NULL::TEXT AS llp2_tstate
, NULL::TEXT AS llp2_tcondition
, NULL::TEXT AS llp1_tstate
, NULL::TEXT AS llp1_tcondition
, NULL::TEXT AS llc_tstate
, NULL::TEXT AS llc_tcondition
, NULL::TEXT AS lli2_tstate
, NULL::TEXT AS lli2_tcondition
, NULL::TEXT AS lli1_tstate
, NULL::TEXT AS lli1_tcondition
, NULL::TEXT AS rli1_tstate
, NULL::TEXT AS rli1_tcondition
, NULL::TEXT AS rli2_tstate
, NULL::TEXT AS rli2_tcondition
, NULL::TEXT AS rlc_tstate
, NULL::TEXT AS rlc_tcondition
, NULL::TEXT AS rlp1_tstate
, NULL::TEXT AS rlp1_tcondition
, NULL::TEXT AS rlp2_tstate
, NULL::TEXT AS rlp2_tcondition
, NULL::TEXT AS rlm1_tstate
, NULL::TEXT AS rlm1_tcondition
, NULL::TEXT AS rlm2_tstate
, NULL::TEXT AS rlm2_tcondition
, NULL::TEXT AS rlm3_tstate
, NULL::TEXT AS rlm3_tcondition
, NULL::TEXT AS drum2_tstate
, NULL::TEXT AS drum2_tcondition
, NULL::TEXT AS drum1_tstate
, NULL::TEXT AS drum1_tcondition
, NULL::TEXT AS druc_tstate
, NULL::TEXT AS druc_tcondition
, NULL::TEXT AS drui2_tstate
, NULL::TEXT AS drui2_tcondition
, NULL::TEXT AS drui1_tstate
, NULL::TEXT AS drui1_tcondition
, NULL::TEXT AS dlui1_tstate
, NULL::TEXT AS dlui1_tcondition
, NULL::TEXT AS dlui2_tstate
, NULL::TEXT AS dlui2_tcondition
, NULL::TEXT AS dluc_tstate
, NULL::TEXT AS dluc_tcondition
, NULL::TEXT AS dlum1_tstate
, NULL::TEXT AS dlum1_tcondition
, NULL::TEXT AS dlum2_tstate
, NULL::TEXT AS dlum2_tcondition
, NULL::TEXT AS dllm2_tstate
, NULL::TEXT AS dllm2_tcondition
, NULL::TEXT AS dllm1_tstate
, NULL::TEXT AS dllm1_tcondition
, NULL::TEXT AS dllc_tstate
, NULL::TEXT AS dllc_tcondition
, NULL::TEXT AS dlli2_tstate
, NULL::TEXT AS dlli2_tcondition
, NULL::TEXT AS dlli1_tstate
, NULL::TEXT AS dlli1_tcondition
, NULL::TEXT AS drli1_tstate
, NULL::TEXT AS drli1_tcondition
, NULL::TEXT AS drli2_tstate
, NULL::TEXT AS drli2_tcondition
, NULL::TEXT AS drlc_tstate
, NULL::TEXT AS drlc_tcondition
, NULL::TEXT AS drlm1_tstate
, NULL::TEXT AS drlm1_tcondition
, NULL::TEXT AS drlm2_tstate
, NULL::TEXT AS drlm2_tcondition
, NULL::TEXT AS teethnotes
, NULL::TEXT AS caninenotes
WHERE _raise_babase_exception(
'Cannot select DART_TEETH_UPLOAD'
|| ': The only use of the DART_TEETH_UPLOAD view is to insert'
|| ' new data into its related tables');
Figure 6.102. Entity Relationship Diagram of the DART_TEETH_UPLOAD View
Table 6.47. Columns in the DART_TEETH_UPLOAD View
| Column | Uploads into | Description |
|---|---|---|
| Name | Nowhere; used to determine this darting's Dartid | The BIOGRAPH.Name associated with this darting's Sname. |
| Sname | Nowhere; used to determine this darting's Dartid | This darting's Sname. |
| Sex | Nowhere; used to determine this darting's Dartid | The BIOGRAPH.Sex associated with this darting's Sname. |
| Dartdate | Nowhere; used to determine this darting's Dartid | The Date of the darting. |
| TOOTHCODE_tstate | TEETH.Tstate | (Multiple columns, one for each Toothcode) The state of the tooth, e.g. present, erupting, missing, etc. |
| TOOTHCODE_tcondition | TEETH.Tcondition | (Multiple columns, one for each Toothcode) The condition of the tooth, e.g. healthy, cracked, etc. |
| Teethnotes | DARTINGS.Teethnotes | General notes about the teeth. |
| Caninenotes | DARTINGS.Caninenotes | General notes about the canines. |
Only INSERT is allowed on DART_TEETH_UPLOAD; SELECT, UPDATE, and DELETE are not allowed. Inserting a row into DART_TEETH_UPLOAD inserts one or more rows into TEETH as described above.
This view is used to insert data about the measured circumference of a darted individual's testes.
Each row inserted into this view represents a single darting. The values in each of the columns may be used to update data in DARTINGS and to insert data into TESTES_ARC. A single row inserted into this view may result in the addition of many rows into TESTES_ARC.
The Name, Sname, Sex, and Dartdate columns are not inserted anywhere. Instead, they are used to identify the darting — that is, to identify the appopriate DARTINGS.Dartid — to which all the row's data belong. As discussed earlier, it is important that the darting event be added to DARTINGS before adding data via this view.
This view includes several sets of three columns, in which each "set" records the data for a single testicle's measurement: a "side" column indicating which testicle is being measured (left or right), a "length" column, and a "width" column. Each of these three-column sets represents a single row that is inserted into TESTES_ARC.
To ensure compatibility with an earlier method of uploading these data, the names of the columns in each set specify a particular side, e.g. the LTestesSide1, LTestesLength1, and LTestesWidth1 columns are implied to only contain data about the left testicle. Regardless of the name of the column, it is the "side" column that truly indicates the side to which the set's data will be attributed.
Tip
If a single darting included more than twelve testicle measurements and you cannot fit them all in a single row of this view, insert another row into this view to insert up to twelve more.
Figure 6.103. Query Defining the DART_TESTES_ARC_UPLOAD View
SELECT NULL::TEXT AS name
, NULL::TEXT AS sname
, NULL::TEXT AS sex
, NULL::TEXT AS dartdate
, NULL::TEXT AS type_descr
, NULL::TEXT AS ltesteslength1
, NULL::TEXT AS ltesteswidth1
, NULL::TEXT AS ltestesside1
, NULL::TEXT AS rtesteslength1
, NULL::TEXT AS rtesteswidth1
, NULL::TEXT AS rtestesside1
, NULL::TEXT AS ltesteslength2
, NULL::TEXT AS ltesteswidth2
, NULL::TEXT AS ltestesside2
, NULL::TEXT AS rtesteslength2
, NULL::TEXT AS rtesteswidth2
, NULL::TEXT AS rtestesside2
, NULL::TEXT AS ltesteslength3
, NULL::TEXT AS ltesteswidth3
, NULL::TEXT AS ltestesside3
, NULL::TEXT AS rtesteslength3
, NULL::TEXT AS rtesteswidth3
, NULL::TEXT AS rtestesside3
, NULL::TEXT AS ltesteslength4
, NULL::TEXT AS ltesteswidth4
, NULL::TEXT AS ltestesside4
, NULL::TEXT AS rtesteslength4
, NULL::TEXT AS rtesteswidth4
, NULL::TEXT AS rtestesside4
, NULL::TEXT AS ltesteslength5
, NULL::TEXT AS ltesteswidth5
, NULL::TEXT AS ltestesside5
, NULL::TEXT AS rtesteslength5
, NULL::TEXT AS rtesteswidth5
, NULL::TEXT AS rtestesside5
, NULL::TEXT AS ltesteslength6
, NULL::TEXT AS ltesteswidth6
, NULL::TEXT AS ltestesside6
, NULL::TEXT AS rtesteslength6
, NULL::TEXT AS rtesteswidth6
, NULL::TEXT AS rtestesside6
, NULL::TEXT AS testesnotes
WHERE _raise_babase_exception(
'Cannot select DART_TESTES_ARC_UPLOAD'
|| ': The only use of the DART_TESTES_ARC_UPLOAD view is to insert'
|| ' new data into its related tables');
Figure 6.104. Entity Relationship Diagram of the DART_TESTES_ARC_UPLOAD View
Table 6.48. Columns in the DART_TESTES_ARC_UPLOAD View
| Column | Uploads into | Description |
|---|---|---|
| Name | Nowhere; used to determine this darting's Dartid | The BIOGRAPH.Name associated with this darting's Sname. |
| Sname | Nowhere; used to determine this darting's Dartid | This darting's Sname. |
| Sex | Nowhere; used to determine this darting's Dartid | The BIOGRAPH.Sex associated with this darting's Sname. |
| Dartdate | Nowhere; used to determine this darting's Dartid | The Date of the darting. |
| LTestesSideN | TESTES_ARC.Testside | Indication of left or right testicle. It is
presumed but not required that a value of
L be supplied, indicating the
length and width are of the left testicle. |
| LTestesLengthN | TESTES_ARC.Testlength | The measured length of the testicle. |
| LTestesWidthN | TESTES_ARC.Testwidth | The measured width of the testicle. |
| RTestesSideN | TESTES_ARC.Testside | Indication of left or right testicle. It is
presumed but not required that a value of
R be supplied, indicating the
length and width are of the right testicle. |
| RTestesLengthN | TESTES_ARC.Testlength | The measured length of the testicle. |
| RTestesWidthN | TESTES_ARC.Testwidth | The measured width of the testicle. |
| Other_Notes_Measures | DARTINGS.Testesnotes | Notes regarding any of these testicle measurements. |
Only INSERT is allowed on DART_TESTES_ARC_UPLOAD; SELECT, UPDATE, and DELETE are not allowed. Inserting a row into DART_TESTES_ARC_UPLOAD inserts rows into TESTES_ARC as described above.
This view is used to insert data about the measured diameter of a darted individual's testes.
Each row inserted into this view represents a single darting. The values in each of the columns may be used to update data in DARTINGS and to insert data into TESTES_DIAM. A single row inserted into this view may result in the addition of many rows into TESTES_DIAM.
The Name, Sname, Sex, and Dartdate columns are not inserted anywhere. Instead, they are used to identify the darting — that is, to identify the appopriate DARTINGS.Dartid — to which all the row's data belong. As discussed earlier, it is important that the darting event be added to DARTINGS before adding data via this view.
This view includes several sets of three columns, in which each "set" records the data for a single testicle's measurement: a "side" column indicating which testicle is being measured (left or right), a "length" column, and a "width" column. Each of these three-column sets represents a single row that is inserted into TESTES_DIAM.
To ensure compatibility with an earlier method of uploading these data, the names of the columns in each set specify a particular side, e.g. the LTestesSide1, LTestesLength1, and LTestesWidth1 columns are implied to only contain data about the left testicle. Regardless of the name of the column, it is the "side" column that truly indicates the side to which the set's data will be attributed.
Tip
If a single darting included more than twelve testicle measurements and you cannot fit them all in a single row of this view, insert another row into this view to insert up to twelve more.
Figure 6.105. Query Defining the DART_TESTES_DIAM_UPLOAD View
SELECT NULL::TEXT AS name
, NULL::TEXT AS sname
, NULL::TEXT AS sex
, NULL::TEXT AS dartdate
, NULL::TEXT AS type_descr
, NULL::TEXT AS ltesteslength1
, NULL::TEXT AS ltesteswidth1
, NULL::TEXT AS ltestesside1
, NULL::TEXT AS rtesteslength1
, NULL::TEXT AS rtesteswidth1
, NULL::TEXT AS rtestesside1
, NULL::TEXT AS ltesteslength2
, NULL::TEXT AS ltesteswidth2
, NULL::TEXT AS ltestesside2
, NULL::TEXT AS rtesteslength2
, NULL::TEXT AS rtesteswidth2
, NULL::TEXT AS rtestesside2
, NULL::TEXT AS ltesteslength3
, NULL::TEXT AS ltesteswidth3
, NULL::TEXT AS ltestesside3
, NULL::TEXT AS rtesteslength3
, NULL::TEXT AS rtesteswidth3
, NULL::TEXT AS rtestesside3
, NULL::TEXT AS ltesteslength4
, NULL::TEXT AS ltesteswidth4
, NULL::TEXT AS ltestesside4
, NULL::TEXT AS rtesteslength4
, NULL::TEXT AS rtesteswidth4
, NULL::TEXT AS rtestesside4
, NULL::TEXT AS ltesteslength5
, NULL::TEXT AS ltesteswidth5
, NULL::TEXT AS ltestesside5
, NULL::TEXT AS rtesteslength5
, NULL::TEXT AS rtesteswidth5
, NULL::TEXT AS rtestesside5
, NULL::TEXT AS ltesteslength6
, NULL::TEXT AS ltesteswidth6
, NULL::TEXT AS ltestesside6
, NULL::TEXT AS rtesteslength6
, NULL::TEXT AS rtesteswidth6
, NULL::TEXT AS rtestesside6
, NULL::TEXT AS testesnotes
WHERE _raise_babase_exception(
'Cannot select DART_TESTES_DIAM_UPLOAD'
|| ': The only use of the DART_TESTES_DIAM_UPLOAD view is to insert'
|| ' new data into its related tables');
Figure 6.106. Entity Relationship Diagram of the DART_TESTES_DIAM_UPLOAD View
Table 6.49. Columns in the DART_TESTES_DIAM_UPLOAD View
| Column | Uploads into | Description |
|---|---|---|
| Name | Nowhere; used to determine this darting's Dartid | The BIOGRAPH.Name associated with this darting's Sname. |
| Sname | Nowhere; used to determine this darting's Dartid | This darting's Sname. |
| Sex | Nowhere; used to determine this darting's Dartid | The BIOGRAPH.Sex associated with this darting's Sname. |
| Dartdate | Nowhere; used to determine this darting's Dartid | The Date of the darting. |
| LTestesSideN | TESTES_DIAM.Testside | Indication of left or right testicle. It is
presumed but not required that a value of
L be supplied, indicating the
length and width are of the left testicle. |
| LTestesLengthN | TESTES_DIAM.Testlength | The measured length of the testicle. |
| LTestesWidthN | TESTES_DIAM.Testwidth | The measured width of the testicle. |
| RTestesSideN | TESTES_DIAM.Testside | Indication of left or right testicle. It is
presumed but not required that a value of
R be supplied, indicating the
length and width are of the right testicle. |
| RTestesLengthN | TESTES_DIAM.Testlength | The measured length of the testicle. |
| RTestesWidthN | TESTES_DIAM.Testwidth | The measured width of the testicle. |
| Other_Notes_Measures | DARTINGS.Testesnotes | Notes regarding any of these testicle measurements. |
Only INSERT is allowed on DART_TESTES_DIAM_UPLOAD; SELECT, UPDATE, and DELETE are not allowed. Inserting a row into DART_TESTES_DIAM_UPLOAD inserts rows into TESTES_DIAM as described above.
This view is used to insert data about the parasite infestation (or lack thereof) of a darted individual.
Each row inserted into this view represents the parasite count of a specific body part, counted during the indicated darting. The values in each of the columns may be used to update data in DARTINGS but are mostly used to insert data into the TICKS table. A single row inserted into this view corresponds to the addition of a single row into TICKS.
The Name, Sname, Sex, and Dartdate columns are not inserted anywhere. Instead, they are used to identify the darting — that is, to identify the appopriate DARTINGS.Dartid — to which all the row's data belong. As discussed earlier, it is important that the darting event be added to DARTINGS before adding data via this view.
This view will not overwrite data in DARTINGS.Ticknotes. If
a row has a non-NULL Other_Notes_Measures, an error will be
returned if the darting's Ticknotes
is already non-NULL. This may help prevent the addition of
duplicate data.
Caution
Because much of the darting data can involve collection of multiple sets of repeated data per darting, there are few checks which can prevent duplicate data.
For example, there are no restrictions which require that all the data which pertain to a given darting be recorded in contiguous rows so repetition of a darting in a later part of an uploaded file is not detected. Care must be taken not to upload the same data twice.
Figure 6.107. Query Defining the DART_TICKS_UPLOAD View
SELECT NULL::TEXT AS name
, NULL::TEXT AS sname
, NULL::TEXT AS sex
, NULL::TEXT AS dartdate
, NULL::TEXT AS pcount
, NULL::TEXT AS bodypart
, NULL::TEXT AS pkind
, NULL::TEXT AS pstatus
, NULL::TEXT AS pnotes
, NULL::TEXT AS other_notes_measures
WHERE _raise_babase_exception(
'Cannot select DART_TICKS_UPLOAD'
|| ': The only use of the DART_TICKS_UPLOAD view is to insert'
|| ' new data into its related tables');
Figure 6.108. Entity Relationship Diagram of the DART_TICKS_UPLOAD View
Table 6.50. Columns in the DART_TICKS_UPLOAD View
| Column | Uploads into | Description |
|---|---|---|
| Name | Nowhere; used to determine this darting's Dartid | The BIOGRAPH.Name associated with this darting's Sname. |
| Sname | Nowhere; used to determine this darting's Dartid | This darting's Sname. |
| Sex | Nowhere; used to determine this darting's Dartid | The BIOGRAPH.Sex associated with this darting's Sname. |
| Dartdate | Nowhere; used to determine this darting's Dartid | The Date of the darting. |
| PCount | TICKS.Tickcount | The number of parasites found on the designated body part. |
| Bodypart | TICKS.Bodypart | The body part examined for parasites. |
| Pkind | TICKS.Tickkind | The kind of parasite counted. |
| Pstatus | TICKS.Tickstatus | The classification of the count itself. |
| Pnotes | TICKS.Tickbpnotes | Notes on this count of this parasite type on this body part. |
| Other_Notes_Measures | DARTINGS.Ticknotes | General notes on the counting of ticks and other parasites. |
Only INSERT is allowed on DART_TICKS_UPLOAD; SELECT, UPDATE, and DELETE are not allowed. Inserting a row into DART_TICKS_UPLOAD inserts rows into TICKS as described above. It may also update a row in DARTINGS, as described below.
When the inserted row has a non-NULL
Other_Notes_Measures, the related DARTINGS.Ticknotes
for this darting is updated with that value.
Caution
When multiple rows from the same darting each have different Other_Notes_Measures values, each row's Other_Notes_Measures value replaces the note from the previous row. Thus, if a single darting has comments/notes from multiple body parts or parasite types, those comments should all be collated into the Other_Notes_Measures of a single row before insertion.
This view is used to insert data about the vaginal pH of a darted individual.
Each row inserted into this view represents a single pH measurement performed during a darting, that is, a single row in the VAGINAL_PHS table.
The Name, Sname, Sex, and Dartdate columns are not inserted anywhere. Instead, they are used to identify the darting — that is, to identify the appopriate DARTINGS.Dartid — to which all the row's data belong. As discussed earlier, it is important that the darting event be added to DARTINGS before adding data via this view.
Figure 6.109. Query Defining the DART_VAGINAL_PHS_UPLOAD View
SELECT NULL::TEXT AS name
, NULL::TEXT AS sname
, NULL::TEXT AS sex
, NULL::TEXT AS dartdate
, NULL::TEXT AS ph
WHERE _raise_babase_exception(
'Cannot select DART_VAGINAL_PHS_UPLOAD'
|| ': The only use of the DART_VAGINAL_PHS_UPLOAD view is to insert'
|| ' new data into its related tables');
Figure 6.110. Entity Relationship Diagram of the DART_VAGINAL_PHS_UPLOAD View
Table 6.51. Columns in the DART_VAGINAL_PHS_UPLOAD View
| Column | Uploads into | Description |
|---|---|---|
| Name | Nowhere; used to determine this darting's Dartid | The BIOGRAPH.Name associated with this darting's Sname. |
| Sname | Nowhere; used to determine this darting's Dartid | This darting's Sname. |
| Sex | Nowhere; used to determine this darting's Dartid | The BIOGRAPH.Sex associated with this darting's Sname. |
| Dartdate | Nowhere; used to determine this darting's Dartid | The Date of the darting. |
| PH | VAGINAL_PHS.PH | The vaginal pH measurement. |
Only INSERT is allowed on DART_VAGINAL_PHS_UPLOAD; SELECT, UPDATE, and DELETE are not allowed. Inserting a row into DART_VAGINAL_PHS_UPLOAD inserts a row into VAGINAL_PHS as described above.
This view is used to insert data about white blood cell counts performed on blood smears that were collected during a darting.
Each row inserted into this view represents a single count of a blood smear. That is, a single row in the WBC_COUNTS table.
The Name, Sname, Sex, and Dartdate columns are not inserted anywhere. Instead, they are used to identify the darting — that is, to identify the appopriate DARTINGS.Dartid — to which all the row's data belong. As discussed earlier, it is important that the darting event be added to DARTINGS before adding data via this view.
Figure 6.111. Query Defining the DART_WBC_COUNTS_UPLOAD View
SELECT NULL::TEXT AS name
, NULL::TEXT AS sname
, NULL::TEXT AS sex
, NULL::TEXT AS dartdate
, NULL::TEXT AS count_date
, NULL::TEXT AS basophils
, NULL::TEXT AS eosinophils
, NULL::TEXT AS monocytes
, NULL::TEXT AS lymphocytes
, NULL::TEXT AS neutrophils
, NULL::TEXT AS counted_by
, NULL::TEXT AS slide_number
, NULL::TEXT AS comments
WHERE _raise_babase_exception(
'Cannot select DART_WBC_COUNTS_UPLOAD'
|| ': The only use of the DART_WBC_COUNTS_UPLOAD view is to insert'
|| ' new data into the WBC_COUNTS table');
Figure 6.112. Entity Relationship Diagram of the DART_WBC_COUNTS_UPLOAD View
Table 6.52. Columns in the DART_WBC_COUNTS_UPLOAD View
| Column | Uploads into | Description |
|---|---|---|
| Name | Nowhere; used to determine this darting's Dartid | The BIOGRAPH.Name associated with this darting's Sname. |
| Sname | Nowhere; used to determine this darting's Dartid | This darting's Sname. |
| Sex | Nowhere; used to determine this darting's Dartid | The BIOGRAPH.Sex associated with this darting's Sname. |
| Dartdate | Nowhere; used to determine this darting's Dartid | The Date of the darting. |
| Count_Date | WBC_COUNTS.Count_Date | The date that the blood smear was counted. |
| Basophils | WBC_COUNTS.Basophils | The number of basophils counted. |
| Eosinophils | WBC_COUNTS.Eosinophils | The number of eosinophils counted. |
| Monocytes | WBC_COUNTS.Monocytes | The number of monocytes counted. |
| Lymphocytes | WBC_COUNTS.Lymphocytes | The number of lymphocytes counted. |
| Neutrophils | WBC_COUNTS.Neutrophils | The number of neutrophils counted. |
| Counted_By | WBC_COUNTS.Counted_By | The initials of the person who performed this count. |
| Slide_number | WBC_COUNTS.Slide_number | An integer indicating which of this darting's blood smear slides was counted for this row.. |
| Comments | WBC_COUNTS.Comments | Comments or miscellaneous information about the counts on this slide. |
Only INSERT is allowed on DART_WBC_COUNTS_UPLOAD; SELECT, UPDATE, and DELETE are not allowed. Inserting a row into DART_WBC_COUNTS_UPLOAD inserts a row into WBC_COUNTS as described above.
Contains one row for every darting.[269] Each row contains columns from DART_SAMPLES for every existing DART_SAMPLES.DS_Type. This shows all samples
collected during the given darting in one row. When there is
no information about how many of a particular DS_Type were collected, the column
indicating that sample type is NULL.
One column appears in DSAMPLES for each DART_SAMPLE_TYPES.DS_Type.
Figure 6.113. Query Defining the DSAMPLES View
SELECT dartings.dartid
, dartings.sname
, dartings.date
, members.grp
, blood_unspecs.num AS bloodunspec
, blood_paxgenes.num AS bloodpaxgene
, blood_purpletops.num AS bloodpurpletops
, blood_separators.num AS bloodseptube
, blood_cpts.num AS bloodcpt
, blood_trucultures.num AS bloodtruculture
, blood_smears.num AS bloodsmear
, tc_bloods.num AS tcblood
, hair_unspecs.num AS hairunspec
, hair_lengths.num AS hairlength
, hair_cu_zns.num AS haircu_zn
, teeth_3mouths.num AS mouthphotos3
, teeth_lmandmolds.num AS lmandmold
, teeth_lmaxmolds.num AS lmaxillamold
, teeth_lmol1mol2s.num AS lm1m2siliconemold
, skin_punchs.num AS skinpunch
, tc_skins.num AS tcskin
, vag_swabs.num AS vaginalswab
, cerv_swabs.num AS cervicalswab
, fecal_formalin.num AS fecal_formalin
, palm_swab.num AS palm_swab
, tongue_swab.num AS tongue_swab
, tooth_plaque_swab.num as tooth_plaque_swab
, vagswab_microbiome.num AS vagswab_microbiome
, glans_penis_swab.num AS glans_penis_swab
, fecal_microbiome.num AS fecal_microbiome
, nostrils_swab.num AS nostrils_swab
, skin_behind_ear_swab.num AS skin_behind_ear_swab
, skin_inside_elbow_swab.num AS skin_inside_elbow_swab
FROM dartings
JOIN members
ON dartings.sname = members.sname
AND dartings.date = members.date
LEFT JOIN dart_samples blood_unspecs
ON dartings.dartid = blood_unspecs.dartid
AND blood_unspecs.ds_type = 1
LEFT JOIN dart_samples blood_paxgenes
ON dartings.dartid = blood_paxgenes.dartid
AND blood_paxgenes.ds_type = 2
LEFT JOIN dart_samples blood_purpletops
ON dartings.dartid = blood_purpletops.dartid
AND blood_purpletops.ds_type = 3
LEFT JOIN dart_samples blood_separators
ON dartings.dartid = blood_separators.dartid
AND blood_separators.ds_type = 4
LEFT JOIN dart_samples blood_cpts
ON dartings.dartid = blood_cpts.dartid
AND blood_cpts.ds_type = 5
LEFT JOIN dart_samples blood_trucultures
ON dartings.dartid = blood_trucultures.dartid
AND blood_trucultures.ds_type = 6
LEFT JOIN dart_samples blood_smears
ON dartings.dartid = blood_smears.dartid
AND blood_smears.ds_type = 7
LEFT JOIN dart_samples hair_unspecs
ON dartings.dartid = hair_unspecs.dartid
AND hair_unspecs.ds_type = 8
LEFT JOIN dart_samples hair_lengths
ON dartings.dartid = hair_lengths.dartid
AND hair_lengths.ds_type = 9
LEFT JOIN dart_samples hair_cu_zns
ON dartings.dartid = hair_cu_zns.dartid
AND hair_cu_zns.ds_type = 10
LEFT JOIN dart_samples teeth_3mouths
ON dartings.dartid = teeth_3mouths.dartid
AND teeth_3mouths.ds_type = 11
LEFT JOIN dart_samples teeth_lmandmolds
ON dartings.dartid = teeth_lmandmolds.dartid
AND teeth_lmandmolds.ds_type = 12
LEFT JOIN dart_samples teeth_lmaxmolds
ON dartings.dartid = teeth_lmaxmolds.dartid
AND teeth_lmaxmolds.ds_type = 13
LEFT JOIN dart_samples teeth_lmol1mol2s
ON dartings.dartid = teeth_lmol1mol2s.dartid
AND teeth_lmol1mol2s.ds_type = 14
LEFT JOIN dart_samples skin_punchs
ON dartings.dartid = skin_punchs.dartid
AND skin_punchs.ds_type = 15
LEFT JOIN dart_samples vag_swabs
ON dartings.dartid = vag_swabs.dartid
AND vag_swabs.ds_type = 16
LEFT JOIN dart_samples cerv_swabs
ON dartings.dartid = cerv_swabs.dartid
AND cerv_swabs.ds_type = 17
LEFT JOIN dart_samples tc_bloods
ON dartings.dartid = tc_bloods.dartid
AND tc_bloods.ds_type = 18
LEFT JOIN dart_samples tc_skins
ON dartings.dartid = tc_skins.dartid
AND tc_skins.ds_type = 19
LEFT JOIN dart_samples fecal_formalin
ON dartings.dartid = fecal_formalin.dartid
AND fecal_formalin.ds_type = 20
LEFT JOIN dart_samples palm_swab
ON dartings.dartid = palm_swab.dartid
AND palm_swab.ds_type = 22
LEFT JOIN dart_samples tongue_swab
ON dartings.dartid = tongue_swab.dartid
AND tongue_swab.ds_type = 23
LEFT JOIN dart_samples tooth_plaque_swab
ON dartings.dartid = tooth_plaque_swab.dartid
AND tooth_plaque_swab.ds_type = 24
LEFT JOIN dart_samples vagswab_microbiome
ON dartings.dartid = vagswab_microbiome.dartid
AND vagswab_microbiome.ds_type = 25
LEFT JOIN dart_samples glans_penis_swab
ON dartings.dartid = glans_penis_swab.dartid
AND glans_penis_swab.ds_type = 26
LEFT JOIN dart_samples fecal_microbiome
ON dartings.dartid = fecal_microbiome.dartid
AND fecal_microbiome.ds_type = 27
LEFT JOIN dart_samples nostrils_swab
ON dartings.dartid = nostrils_swab.dartid
AND nostrils_swab.ds_type = 28
LEFT JOIN dart_samples skin_behind_ear_swab
ON dartings.dartid = skin_behind_ear_swab.dartid
AND skin_behind_ear_swab.ds_type = 29
LEFT JOIN dart_samples skin_inside_elbow_swab
ON dartings.dartid = skin_inside_elbow_swab.dartid
AND skin_inside_elbow_swab.ds_type = 30;
Because most of the columns in DSAMPLES are based on the rows present in DART_SAMPLE_TYPES there is not a description of
each column here. For columns indicating a number of a
number of collected samples, the column name is always an
abbreviated version of the DS_Type description. For
example, a DART_SAMPLE_TYPES.DS_Type whose DART_SAMPLE_TYPES.Descr is LEFT
MANDIBLE MOLD will be counted in the
DSAMPLES.Lmandmold column. These columns are described below
in a generic fashion.
Table 6.53. Columns in the DSAMPLES View
| Column | From | Description |
|---|---|---|
| Dartid | DARTINGS.Dartid | Identifier of the darting event. |
| Sname | DARTINGS.Sname | The Sname of the darted individual. |
| Date | DARTINGS.Date | The date of the darting. |
| Grp | MEMBERS.Grp | The study group the individual was in, on the darting date. |
| [Sample counts] | DART_SAMPLES.Num | The number of samples collected of the type indicated by the column name. |
Contains one row for every darting during which
dentition information was taken.[270] Each row contains columns from TEETH for every existing TOOTHCODES.Toothcode
value. This shows all the tooth-related information
collecting during the given darting as one row, in a fashion
that is structured based on the teeth found in baboons.
When there is no information on a particular tooth the
values in the columns having to do with that tooth are
NULL.
Two columns appear in DENT_CODES for every TOOTHCODES.Toothcode
value. A column named TCtstate, where the
TOOTHCODES.Toothcode
value replaces the letters “TC”, shows the TEETH.Tstate of the tooth.
A column named TCtcondition, where the
TOOTHCODES.Toothcode
value replaces the letters “TC”, shows the TEETH.Tcondition of the
tooth.
Warning
Adding or deleting TOOTHCODES.Toothcode does not automatically change the DENT_CODES view. The view must be manually re-coded to reflect changes made to TOOTHCODES.
This view is useful when joined with the DARTINGS table on Dartid.
Figure 6.114. Query Defining the DENT_CODES View
SELECT teethdartids.dartid AS dartid
, rum3.rum3tstate AS rum3tstate
, rum3.rum3tcondition AS rum3tcondition
, rum2.rum2tstate AS rum2tstate
, rum2.rum2tcondition AS rum2tcondition
, rum1.rum1tstate AS rum1tstate
, rum1.rum1tcondition AS rum1tcondition
, rup2.rup2tstate AS rup2tstate
, rup2.rup2tcondition AS rup2tcondition
, rup1.rup1tstate AS rup1tstate
, rup1.rup1tcondition AS rup1tcondition
, ruc.ructstate AS ructstate
, ruc.ructcondition AS ructcondition
, rui2.rui2tstate AS rui2tstate
, rui2.rui2tcondition AS rui2tcondition
, rui1.rui1tstate AS rui1tstate
, rui1.rui1tcondition AS rui1tcondition
, lui1.lui1tstate AS lui1tstate
, lui1.lui1tcondition AS lui1tcondition
, lui2.lui2tstate AS lui2tstate
, lui2.lui2tcondition AS lui2tcondition
, luc.luctstate AS luctstate
, luc.luctcondition AS luctcondition
, lup1.lup1tstate AS lup1tstate
, lup1.lup1tcondition AS lup1tcondition
, lup2.lup2tstate AS lup2tstate
, lup2.lup2tcondition AS lup2tcondition
, lum1.lum1tstate AS lum1tstate
, lum1.lum1tcondition AS lum1tcondition
, lum2.lum2tstate AS lum2tstate
, lum2.lum2tcondition AS lum2tcondition
, lum3.lum3tstate AS lum3tstate
, lum3.lum3tcondition AS lum3tcondition
, llm3.llm3tstate AS llm3tstate
, llm3.llm3tcondition AS llm3tcondition
, llm2.llm2tstate AS llm2tstate
, llm2.llm2tcondition AS llm2tcondition
, llm1.llm1tstate AS llm1tstate
, llm1.llm1tcondition AS llm1tcondition
, llp2.llp2tstate AS llp2tstate
, llp2.llp2tcondition AS llp2tcondition
, llp1.llp1tstate AS llp1tstate
, llp1.llp1tcondition AS llp1tcondition
, llc.llctstate AS llctstate
, llc.llctcondition AS llctcondition
, lli2.lli2tstate AS lli2tstate
, lli2.lli2tcondition AS lli2tcondition
, lli1.lli1tstate AS lli1tstate
, lli1.lli1tcondition AS lli1tcondition
, rli1.rli1tstate AS rli1tstate
, rli1.rli1tcondition AS rli1tcondition
, rli2.rli2tstate AS rli2tstate
, rli2.rli2tcondition AS rli2tcondition
, rlc.rlctstate AS rlctstate
, rlc.rlctcondition AS rlctcondition
, rlp1.rlp1tstate AS rlp1tstate
, rlp1.rlp1tcondition AS rlp1tcondition
, rlp2.rlp2tstate AS rlp2tstate
, rlp2.rlp2tcondition AS rlp2tcondition
, rlm1.rlm1tstate AS rlm1tstate
, rlm1.rlm1tcondition AS rlm1tcondition
, rlm2.rlm2tstate AS rlm2tstate
, rlm2.rlm2tcondition AS rlm2tcondition
, rlm3.rlm3tstate AS rlm3tstate
, rlm3.rlm3tcondition AS rlm3tcondition
, drum2.drum2tstate AS drum2tstate
, drum2.drum2tcondition AS drum2tcondition
, drum1.drum1tstate AS drum1tstate
, drum1.drum1tcondition AS drum1tcondition
, druc.dructstate AS dructstate
, druc.dructcondition AS dructcondition
, drui2.drui2tstate AS drui2tstate
, drui2.drui2tcondition AS drui2tcondition
, drui1.drui1tstate AS drui1tstate
, drui1.drui1tcondition AS drui1tcondition
, dlui1.dlui1tstate AS dlui1tstate
, dlui1.dlui1tcondition AS dlui1tcondition
, dlui2.dlui2tstate AS dlui2tstate
, dlui2.dlui2tcondition AS dlui2tcondition
, dluc.dluctstate AS dluctstate
, dluc.dluctcondition AS dluctcondition
, dlum1.dlum1tstate AS dlum1tstate
, dlum1.dlum1tcondition AS dlum1tcondition
, dlum2.dlum2tstate AS dlum2tstate
, dlum2.dlum2tcondition AS dlum2tcondition
, dllm2.dllm2tstate AS dllm2tstate
, dllm2.dllm2tcondition AS dllm2tcondition
, dllm1.dllm1tstate AS dllm1tstate
, dllm1.dllm1tcondition AS dllm1tcondition
, dllc.dllctstate AS dllctstate
, dllc.dllctcondition AS dllctcondition
, dlli2.dlli2tstate AS dlli2tstate
, dlli2.dlli2tcondition AS dlli2tcondition
, dlli1.dlli1tstate AS dlli1tstate
, dlli1.dlli1tcondition AS dlli1tcondition
, drli1.drli1tstate AS drli1tstate
, drli1.drli1tcondition AS drli1tcondition
, drli2.drli2tstate AS drli2tstate
, drli2.drli2tcondition AS drli2tcondition
, drlc.drlctstate AS drlctstate
, drlc.drlctcondition AS drlctcondition
, drlm1.drlm1tstate AS drlm1tstate
, drlm1.drlm1tcondition AS drlm1tcondition
, drlm2.drlm2tstate AS drlm2tstate
, drlm2.drlm2tcondition AS drlm2tcondition
FROM (SELECT teeth.dartid
FROM teeth
GROUP BY teeth.dartid)
AS teethdartids
LEFT OUTER JOIN
(SELECT teeth.dartid AS rum3dartid
, teeth.tstate AS rum3tstate
, teeth.tcondition AS rum3tcondition
FROM teeth
WHERE teeth.tooth = 'rum3')
AS rum3
ON rum3.rum3dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rum2dartid
, teeth.tstate AS rum2tstate
, teeth.tcondition AS rum2tcondition
FROM teeth
WHERE teeth.tooth = 'rum2')
AS rum2
ON rum2.rum2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rum1dartid
, teeth.tstate AS rum1tstate
, teeth.tcondition AS rum1tcondition
FROM teeth
WHERE teeth.tooth = 'rum1')
AS rum1
ON rum1.rum1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rup2dartid
, teeth.tstate AS rup2tstate
, teeth.tcondition AS rup2tcondition
FROM teeth
WHERE teeth.tooth = 'rup2')
AS rup2
ON rup2.rup2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rup1dartid
, teeth.tstate AS rup1tstate
, teeth.tcondition AS rup1tcondition
FROM teeth
WHERE teeth.tooth = 'rup1')
AS rup1
ON rup1.rup1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rucdartid
, teeth.tstate AS ructstate
, teeth.tcondition AS ructcondition
FROM teeth
WHERE teeth.tooth = 'ruc')
AS ruc
ON ruc.rucdartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rui2dartid
, teeth.tstate AS rui2tstate
, teeth.tcondition AS rui2tcondition
FROM teeth
WHERE teeth.tooth = 'rui2')
AS rui2
ON rui2.rui2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rui1dartid
, teeth.tstate AS rui1tstate
, teeth.tcondition AS rui1tcondition
FROM teeth
WHERE teeth.tooth = 'rui1')
AS rui1
ON rui1.rui1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS lui1dartid
, teeth.tstate AS lui1tstate
, teeth.tcondition AS lui1tcondition
FROM teeth
WHERE teeth.tooth = 'lui1')
AS lui1
ON lui1.lui1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS lui2dartid
, teeth.tstate AS lui2tstate
, teeth.tcondition AS lui2tcondition
FROM teeth
WHERE teeth.tooth = 'lui2')
AS lui2
ON lui2.lui2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS lucdartid
, teeth.tstate AS luctstate
, teeth.tcondition AS luctcondition
FROM teeth
WHERE teeth.tooth = 'luc')
AS luc
ON luc.lucdartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS lup1dartid
, teeth.tstate AS lup1tstate
, teeth.tcondition AS lup1tcondition
FROM teeth
WHERE teeth.tooth = 'lup1')
AS lup1
ON lup1.lup1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS lup2dartid
, teeth.tstate AS lup2tstate
, teeth.tcondition AS lup2tcondition
FROM teeth
WHERE teeth.tooth = 'lup2')
AS lup2
ON lup2.lup2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS lum1dartid
, teeth.tstate AS lum1tstate
, teeth.tcondition AS lum1tcondition
FROM teeth
WHERE teeth.tooth = 'lum1')
AS lum1
ON lum1.lum1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS lum2dartid
, teeth.tstate AS lum2tstate
, teeth.tcondition AS lum2tcondition
FROM teeth
WHERE teeth.tooth = 'lum2')
AS lum2
ON lum2.lum2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS lum3dartid
, teeth.tstate AS lum3tstate
, teeth.tcondition AS lum3tcondition
FROM teeth
WHERE teeth.tooth = 'lum3')
AS lum3
ON lum3.lum3dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS llm3dartid
, teeth.tstate AS llm3tstate
, teeth.tcondition AS llm3tcondition
FROM teeth
WHERE teeth.tooth = 'llm3')
AS llm3
ON llm3.llm3dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS llm2dartid
, teeth.tstate AS llm2tstate
, teeth.tcondition AS llm2tcondition
FROM teeth
WHERE teeth.tooth = 'llm2')
AS llm2
ON llm2.llm2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS llm1dartid
, teeth.tstate AS llm1tstate
, teeth.tcondition AS llm1tcondition
FROM teeth
WHERE teeth.tooth = 'llm1')
AS llm1
ON llm1.llm1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS llp2dartid
, teeth.tstate AS llp2tstate
, teeth.tcondition AS llp2tcondition
FROM teeth
WHERE teeth.tooth = 'llp2')
AS llp2
ON llp2.llp2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS llp1dartid
, teeth.tstate AS llp1tstate
, teeth.tcondition AS llp1tcondition
FROM teeth
WHERE teeth.tooth = 'llp1')
AS llp1
ON llp1.llp1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS llcdartid
, teeth.tstate AS llctstate
, teeth.tcondition AS llctcondition
FROM teeth
WHERE teeth.tooth = 'llc')
AS llc
ON llc.llcdartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS lli2dartid
, teeth.tstate AS lli2tstate
, teeth.tcondition AS lli2tcondition
FROM teeth
WHERE teeth.tooth = 'lli2')
AS lli2
ON lli2.lli2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS lli1dartid
, teeth.tstate AS lli1tstate
, teeth.tcondition AS lli1tcondition
FROM teeth
WHERE teeth.tooth = 'lli1')
AS lli1
ON lli1.lli1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rli1dartid
, teeth.tstate AS rli1tstate
, teeth.tcondition AS rli1tcondition
FROM teeth
WHERE teeth.tooth = 'rli1')
AS rli1
ON rli1.rli1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rli2dartid
, teeth.tstate AS rli2tstate
, teeth.tcondition AS rli2tcondition
FROM teeth
WHERE teeth.tooth = 'rli2')
AS rli2
ON rli2.rli2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rlcdartid
, teeth.tstate AS rlctstate
, teeth.tcondition AS rlctcondition
FROM teeth
WHERE teeth.tooth = 'rlc')
AS rlc
ON rlc.rlcdartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rlp1dartid
, teeth.tstate AS rlp1tstate
, teeth.tcondition AS rlp1tcondition
FROM teeth
WHERE teeth.tooth = 'rlp1')
AS rlp1
ON rlp1.rlp1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rlp2dartid
, teeth.tstate AS rlp2tstate
, teeth.tcondition AS rlp2tcondition
FROM teeth
WHERE teeth.tooth = 'rlp2')
AS rlp2
ON rlp2.rlp2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rlm1dartid
, teeth.tstate AS rlm1tstate
, teeth.tcondition AS rlm1tcondition
FROM teeth
WHERE teeth.tooth = 'rlm1')
AS rlm1
ON rlm1.rlm1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rlm2dartid
, teeth.tstate AS rlm2tstate
, teeth.tcondition AS rlm2tcondition
FROM teeth
WHERE teeth.tooth = 'rlm2')
AS rlm2
ON rlm2.rlm2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rlm3dartid
, teeth.tstate AS rlm3tstate
, teeth.tcondition AS rlm3tcondition
FROM teeth
WHERE teeth.tooth = 'rlm3')
AS rlm3
ON rlm3.rlm3dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS drum2dartid
, teeth.tstate AS drum2tstate
, teeth.tcondition AS drum2tcondition
FROM teeth
WHERE teeth.tooth = 'drum2')
AS drum2
ON drum2.drum2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS drum1dartid
, teeth.tstate AS drum1tstate
, teeth.tcondition AS drum1tcondition
FROM teeth
WHERE teeth.tooth = 'drum1')
AS drum1
ON drum1.drum1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS drucdartid
, teeth.tstate AS dructstate
, teeth.tcondition AS dructcondition
FROM teeth
WHERE teeth.tooth = 'druc')
AS druc
ON druc.drucdartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS drui2dartid
, teeth.tstate AS drui2tstate
, teeth.tcondition AS drui2tcondition
FROM teeth
WHERE teeth.tooth = 'drui2')
AS drui2
ON drui2.drui2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS drui1dartid
, teeth.tstate AS drui1tstate
, teeth.tcondition AS drui1tcondition
FROM teeth
WHERE teeth.tooth = 'drui1')
AS drui1
ON drui1.drui1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS dlui1dartid
, teeth.tstate AS dlui1tstate
, teeth.tcondition AS dlui1tcondition
FROM teeth
WHERE teeth.tooth = 'dlui1')
AS dlui1
ON dlui1.dlui1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS dlui2dartid
, teeth.tstate AS dlui2tstate
, teeth.tcondition AS dlui2tcondition
FROM teeth
WHERE teeth.tooth = 'dlui2')
AS dlui2
ON dlui2.dlui2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS dlucdartid
, teeth.tstate AS dluctstate
, teeth.tcondition AS dluctcondition
FROM teeth
WHERE teeth.tooth = 'dluc')
AS dluc
ON dluc.dlucdartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS dlum1dartid
, teeth.tstate AS dlum1tstate
, teeth.tcondition AS dlum1tcondition
FROM teeth
WHERE teeth.tooth = 'dlum1')
AS dlum1
ON dlum1.dlum1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS dlum2dartid
, teeth.tstate AS dlum2tstate
, teeth.tcondition AS dlum2tcondition
FROM teeth
WHERE teeth.tooth = 'dlum2')
AS dlum2
ON dlum2.dlum2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS dllm2dartid
, teeth.tstate AS dllm2tstate
, teeth.tcondition AS dllm2tcondition
FROM teeth
WHERE teeth.tooth = 'dllm2')
AS dllm2
ON dllm2.dllm2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS dllm1dartid
, teeth.tstate AS dllm1tstate
, teeth.tcondition AS dllm1tcondition
FROM teeth
WHERE teeth.tooth = 'dllm1')
AS dllm1
ON dllm1.dllm1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS dllcdartid
, teeth.tstate AS dllctstate
, teeth.tcondition AS dllctcondition
FROM teeth
WHERE teeth.tooth = 'dllc')
AS dllc
ON dllc.dllcdartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS dlli2dartid
, teeth.tstate AS dlli2tstate
, teeth.tcondition AS dlli2tcondition
FROM teeth
WHERE teeth.tooth = 'dlli2')
AS dlli2
ON dlli2.dlli2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS dlli1dartid
, teeth.tstate AS dlli1tstate
, teeth.tcondition AS dlli1tcondition
FROM teeth
WHERE teeth.tooth = 'dlli1')
AS dlli1
ON dlli1.dlli1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS drli1dartid
, teeth.tstate AS drli1tstate
, teeth.tcondition AS drli1tcondition
FROM teeth
WHERE teeth.tooth = 'drli1')
AS drli1
ON drli1.drli1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS drli2dartid
, teeth.tstate AS drli2tstate
, teeth.tcondition AS drli2tcondition
FROM teeth
WHERE teeth.tooth = 'drli2')
AS drli2
ON drli2.drli2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS drlcdartid
, teeth.tstate AS drlctstate
, teeth.tcondition AS drlctcondition
FROM teeth
WHERE teeth.tooth = 'drlc')
AS drlc
ON drlc.drlcdartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS drlm1dartid
, teeth.tstate AS drlm1tstate
, teeth.tcondition AS drlm1tcondition
FROM teeth
WHERE teeth.tooth = 'drlm1')
AS drlm1
ON drlm1.drlm1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS drlm2dartid
, teeth.tstate AS drlm2tstate
, teeth.tcondition AS drlm2tcondition
FROM teeth
WHERE teeth.tooth = 'drlm2')
AS drlm2
ON drlm2.drlm2dartid = teethdartids.dartid;
Because the columns in DENT_CODES are based on the rows present in TOOTHCODES there is not a description of each column here. Instead, the columns based on TOOTHCODES.Toothcode are described below in a generic fashion. Each such column is prefaced here with “TC”, which is replaced by a TOOTHCODES.Toothcode value in the actual column name.
Contains one row for every darting during which
dentition information was taken.[271] Each row contains columns from TEETH and TOOTHCODES tables
for every existing TOOTHCODES.Toothsite value. This shows all the
tooth-related information collecting during the given
darting as one row, in a fashion that is structured around
the position of the teeth within the mouth. When there is
no information on a particular tooth the values in the
columns having to do with that tooth are NULL.
Three columns appear in DENT_SITES for every TOOTHCODES.Toothsite
value. A column named TStstate, where the
letter s followed by the TOOTHCODES.Toothsite
value replaces the letters “TS”[272], shows the TEETH.Tstate of the tooth. A column named
TStcondition, where the letter
s followed by the TOOTHCODES.Toothsite
value replaces the letters “TS”, shows the TEETH.Tcondition of the
tooth. And a column named TSdeciduous,
where the letter s followed by the TOOTHCODES.Toothsite
value replaces the letters “TS”, shows the TOOTHCODES.Deciduous
value for the tooth.
Warning
Adding or deleting TOOTHCODES.Toothcode, or changing the TOOTHCODES.Toothsite does not automatically change the DENT_SITES view. The view must be manually re-coded to reflect changes made to TOOTHCODES.
This view is useful when joined with the DARTINGS table on Dartid.
Figure 6.116. Query Defining the DENT_SITES View
SELECT teethdartids.dartid AS dartid
, s1.s1tstate AS s1tstate
, s1.s1tcondition AS s1tcondition
, s1.s1deciduous AS s1deciduous
, s2.s2tstate AS s2tstate
, s2.s2tcondition AS s2tcondition
, s2.s2deciduous AS s2deciduous
, s3.s3tstate AS s3tstate
, s3.s3tcondition AS s3tcondition
, s3.s3deciduous AS s3deciduous
, s4.s4tstate AS s4tstate
, s4.s4tcondition AS s4tcondition
, s4.s4deciduous AS s4deciduous
, s5.s5tstate AS s5tstate
, s5.s5tcondition AS s5tcondition
, s5.s5deciduous AS s5deciduous
, s6.s6tstate AS s6tstate
, s6.s6tcondition AS s6tcondition
, s6.s6deciduous AS s6deciduous
, s7.s7tstate AS s7tstate
, s7.s7tcondition AS s7tcondition
, s7.s7deciduous AS s7deciduous
, s8.s8tstate AS s8tstate
, s8.s8tcondition AS s8tcondition
, s8.s8deciduous AS s8deciduous
, s9.s9tstate AS s9tstate
, s9.s9tcondition AS s9tcondition
, s9.s9deciduous AS s9deciduous
, s10.s10tstate AS s10tstate
, s10.s10tcondition AS s10tcondition
, s10.s10deciduous AS s10deciduous
, s11.s11tstate AS s11tstate
, s11.s11tcondition AS s11tcondition
, s11.s11deciduous AS s11deciduous
, s12.s12tstate AS s12tstate
, s12.s12tcondition AS s12tcondition
, s12.s12deciduous AS s12deciduous
, s13.s13tstate AS s13tstate
, s13.s13tcondition AS s13tcondition
, s13.s13deciduous AS s13deciduous
, s14.s14tstate AS s14tstate
, s14.s14tcondition AS s14tcondition
, s14.s14deciduous AS s14deciduous
, s15.s15tstate AS s15tstate
, s15.s15tcondition AS s15tcondition
, s15.s15deciduous AS s15deciduous
, s16.s16tstate AS s16tstate
, s16.s16tcondition AS s16tcondition
, s16.s16deciduous AS s16deciduous
, s17.s17tstate AS s17tstate
, s17.s17tcondition AS s17tcondition
, s17.s17deciduous AS s17deciduous
, s18.s18tstate AS s18tstate
, s18.s18tcondition AS s18tcondition
, s18.s18deciduous AS s18deciduous
, s19.s19tstate AS s19tstate
, s19.s19tcondition AS s19tcondition
, s19.s19deciduous AS s19deciduous
, s20.s20tstate AS s20tstate
, s20.s20tcondition AS s20tcondition
, s20.s20deciduous AS s20deciduous
, s21.s21tstate AS s21tstate
, s21.s21tcondition AS s21tcondition
, s21.s21deciduous AS s21deciduous
, s22.s22tstate AS s22tstate
, s22.s22tcondition AS s22tcondition
, s22.s22deciduous AS s22deciduous
, s23.s23tstate AS s23tstate
, s23.s23tcondition AS s23tcondition
, s23.s23deciduous AS s23deciduous
, s24.s24tstate AS s24tstate
, s24.s24tcondition AS s24tcondition
, s24.s24deciduous AS s24deciduous
, s25.s25tstate AS s25tstate
, s25.s25tcondition AS s25tcondition
, s25.s25deciduous AS s25deciduous
, s26.s26tstate AS s26tstate
, s26.s26tcondition AS s26tcondition
, s26.s26deciduous AS s26deciduous
, s27.s27tstate AS s27tstate
, s27.s27tcondition AS s27tcondition
, s27.s27deciduous AS s27deciduous
, s28.s28tstate AS s28tstate
, s28.s28tcondition AS s28tcondition
, s28.s28deciduous AS s28deciduous
, s29.s29tstate AS s29tstate
, s29.s29tcondition AS s29tcondition
, s29.s29deciduous AS s29deciduous
, s30.s30tstate AS s30tstate
, s30.s30tcondition AS s30tcondition
, s30.s30deciduous AS s30deciduous
, s31.s31tstate AS s31tstate
, s31.s31tcondition AS s31tcondition
, s31.s31deciduous AS s31deciduous
, s32.s32tstate AS s32tstate
, s32.s32tcondition AS s32tcondition
, s32.s32deciduous AS s32deciduous
FROM (SELECT teeth.dartid
FROM teeth
GROUP BY teeth.dartid)
AS teethdartids
LEFT OUTER JOIN
(SELECT teeth.dartid AS s1dartid
, teeth.tstate AS s1tstate
, teeth.tcondition AS s1tcondition
, toothcodes.deciduous AS s1deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '1'
AND teeth.tooth = toothcodes.tooth)
AS s1
ON s1.s1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s2dartid
, teeth.tstate AS s2tstate
, teeth.tcondition AS s2tcondition
, toothcodes.deciduous AS s2deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '2'
AND teeth.tooth = toothcodes.tooth)
AS s2
ON s2.s2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s3dartid
, teeth.tstate AS s3tstate
, teeth.tcondition AS s3tcondition
, toothcodes.deciduous AS s3deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '3'
AND teeth.tooth = toothcodes.tooth)
AS s3
ON s3.s3dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s4dartid
, teeth.tstate AS s4tstate
, teeth.tcondition AS s4tcondition
, toothcodes.deciduous AS s4deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '4'
AND teeth.tooth = toothcodes.tooth)
AS s4
ON s4.s4dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s5dartid
, teeth.tstate AS s5tstate
, teeth.tcondition AS s5tcondition
, toothcodes.deciduous AS s5deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '5'
AND teeth.tooth = toothcodes.tooth)
AS s5
ON s5.s5dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s6dartid
, teeth.tstate AS s6tstate
, teeth.tcondition AS s6tcondition
, toothcodes.deciduous AS s6deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '6'
AND teeth.tooth = toothcodes.tooth)
AS s6
ON s6.s6dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s7dartid
, teeth.tstate AS s7tstate
, teeth.tcondition AS s7tcondition
, toothcodes.deciduous AS s7deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '7'
AND teeth.tooth = toothcodes.tooth)
AS s7
ON s7.s7dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s8dartid
, teeth.tstate AS s8tstate
, teeth.tcondition AS s8tcondition
, toothcodes.deciduous AS s8deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '8'
AND teeth.tooth = toothcodes.tooth)
AS s8
ON s8.s8dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s9dartid
, teeth.tstate AS s9tstate
, teeth.tcondition AS s9tcondition
, toothcodes.deciduous AS s9deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '9'
AND teeth.tooth = toothcodes.tooth)
AS s9
ON s9.s9dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s10dartid
, teeth.tstate AS s10tstate
, teeth.tcondition AS s10tcondition
, toothcodes.deciduous AS s10deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '10'
AND teeth.tooth = toothcodes.tooth)
AS s10
ON s10.s10dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s11dartid
, teeth.tstate AS s11tstate
, teeth.tcondition AS s11tcondition
, toothcodes.deciduous AS s11deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '11'
AND teeth.tooth = toothcodes.tooth)
AS s11
ON s11.s11dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s12dartid
, teeth.tstate AS s12tstate
, teeth.tcondition AS s12tcondition
, toothcodes.deciduous AS s12deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '12'
AND teeth.tooth = toothcodes.tooth)
AS s12
ON s12.s12dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s13dartid
, teeth.tstate AS s13tstate
, teeth.tcondition AS s13tcondition
, toothcodes.deciduous AS s13deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '13'
AND teeth.tooth = toothcodes.tooth)
AS s13
ON s13.s13dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s14dartid
, teeth.tstate AS s14tstate
, teeth.tcondition AS s14tcondition
, toothcodes.deciduous AS s14deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '14'
AND teeth.tooth = toothcodes.tooth)
AS s14
ON s14.s14dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s15dartid
, teeth.tstate AS s15tstate
, teeth.tcondition AS s15tcondition
, toothcodes.deciduous AS s15deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '15'
AND teeth.tooth = toothcodes.tooth)
AS s15
ON s15.s15dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s16dartid
, teeth.tstate AS s16tstate
, teeth.tcondition AS s16tcondition
, toothcodes.deciduous AS s16deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '16'
AND teeth.tooth = toothcodes.tooth)
AS s16
ON s16.s16dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s17dartid
, teeth.tstate AS s17tstate
, teeth.tcondition AS s17tcondition
, toothcodes.deciduous AS s17deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '17'
AND teeth.tooth = toothcodes.tooth)
AS s17
ON s17.s17dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s18dartid
, teeth.tstate AS s18tstate
, teeth.tcondition AS s18tcondition
, toothcodes.deciduous AS s18deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '18'
AND teeth.tooth = toothcodes.tooth)
AS s18
ON s18.s18dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s19dartid
, teeth.tstate AS s19tstate
, teeth.tcondition AS s19tcondition
, toothcodes.deciduous AS s19deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '19'
AND teeth.tooth = toothcodes.tooth)
AS s19
ON s19.s19dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s20dartid
, teeth.tstate AS s20tstate
, teeth.tcondition AS s20tcondition
, toothcodes.deciduous AS s20deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '20'
AND teeth.tooth = toothcodes.tooth)
AS s20
ON s20.s20dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s21dartid
, teeth.tstate AS s21tstate
, teeth.tcondition AS s21tcondition
, toothcodes.deciduous AS s21deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '21'
AND teeth.tooth = toothcodes.tooth)
AS s21
ON s21.s21dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s22dartid
, teeth.tstate AS s22tstate
, teeth.tcondition AS s22tcondition
, toothcodes.deciduous AS s22deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '22'
AND teeth.tooth = toothcodes.tooth)
AS s22
ON s22.s22dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s23dartid
, teeth.tstate AS s23tstate
, teeth.tcondition AS s23tcondition
, toothcodes.deciduous AS s23deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '23'
AND teeth.tooth = toothcodes.tooth)
AS s23
ON s23.s23dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s24dartid
, teeth.tstate AS s24tstate
, teeth.tcondition AS s24tcondition
, toothcodes.deciduous AS s24deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '24'
AND teeth.tooth = toothcodes.tooth)
AS s24
ON s24.s24dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s25dartid
, teeth.tstate AS s25tstate
, teeth.tcondition AS s25tcondition
, toothcodes.deciduous AS s25deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '25'
AND teeth.tooth = toothcodes.tooth)
AS s25
ON s25.s25dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s26dartid
, teeth.tstate AS s26tstate
, teeth.tcondition AS s26tcondition
, toothcodes.deciduous AS s26deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '26'
AND teeth.tooth = toothcodes.tooth)
AS s26
ON s26.s26dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s27dartid
, teeth.tstate AS s27tstate
, teeth.tcondition AS s27tcondition
, toothcodes.deciduous AS s27deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '27'
AND teeth.tooth = toothcodes.tooth)
AS s27
ON s27.s27dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s28dartid
, teeth.tstate AS s28tstate
, teeth.tcondition AS s28tcondition
, toothcodes.deciduous AS s28deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '28'
AND teeth.tooth = toothcodes.tooth)
AS s28
ON s28.s28dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s29dartid
, teeth.tstate AS s29tstate
, teeth.tcondition AS s29tcondition
, toothcodes.deciduous AS s29deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '29'
AND teeth.tooth = toothcodes.tooth)
AS s29
ON s29.s29dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s30dartid
, teeth.tstate AS s30tstate
, teeth.tcondition AS s30tcondition
, toothcodes.deciduous AS s30deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '30'
AND teeth.tooth = toothcodes.tooth)
AS s30
ON s30.s30dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s31dartid
, teeth.tstate AS s31tstate
, teeth.tcondition AS s31tcondition
, toothcodes.deciduous AS s31deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '31'
AND teeth.tooth = toothcodes.tooth)
AS s31
ON s31.s31dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s32dartid
, teeth.tstate AS s32tstate
, teeth.tcondition AS s32tcondition
, toothcodes.deciduous AS s32deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '32'
AND teeth.tooth = toothcodes.tooth)
AS s32
ON s32.s32dartid = teethdartids.dartid;
Because the columns in DENT_SITES
are based on the rows present in TOOTHCODES there is not a description of each
column here. Instead, the columns based on TOOTHCODES.Toothsite are described below in a
generic fashion. Each such column is prefaced here with
“TS”, which is replaced by the letter
s followed by a TOOTHCODES.Toothsite value in the actual column
name.
Table 6.55. Columns in the DENT_SITES View
| Column | From | Description |
|---|---|---|
| Dartid | TEETH.Dartid | Identifier of the darting event. |
| TStstate | TEETH.Tstate | Code indicating the degree to which the tooth
exists. When NULL no information on the tooth was
recorded during the darting. |
| TStcondition | TEETH.Tcondition | Code indicating the condition of the tooth. |
| TSdeciduous | TOOTHCODES.Deciduous | True when the tooth is
deciduous, False when it is
not. |
Contains one row for every unique Dartid value in the HUMERUSES table.[273] Each row statistically summarizes the HUMERUSES rows having the common Dartid value.
This view is useful when joined with the DARTINGS table on Dartid.
Figure 6.118. Query Defining the HUMERUS_STATS View
SELECT humeruses.dartid AS dartid
, count(*) AS husamps
, avg(humeruses.hulength) AS hulength_mean
, stddev(humeruses.hulength) AS hulength_stddev
, avg(humeruses.huunadjusted) AS huunadjusted_mean
, stddev(humeruses.huunadjusted) AS huunadjusted_stddev
FROM humeruses
GROUP BY humeruses.dartid;
Table 6.56. Columns in the HUMERUS_STATS View
| Column | From | Description |
|---|---|---|
| Dartid | HUMERUSES.Dartid | Identifier of the darting event. |
| Husamps | Computed | Number of HUMERUSES rows having the given Dartid value -- the number of humerus length measurements taken during the darting. |
| Hulength_mean | HUMERUSES.Hulength (computed) | The arithmetic mean of the humerus length measurements related to the given Dartid -- the mean of the humerus length measurements taken during the darting. |
| Hulength_stddev | HUMERUSES.Hulength (computed) | The standard deviation of the humerus length measurements related to the given Dartid -- the standard deviation of the humerus length measurements taken during the darting. |
| Huunadjusted_mean | HUMERUSES.Huunadjusted (computed) | The arithmetic mean of the unadjusted humerus length measurements related to the given Dartid -- the mean of the unadjusted humerus length measurements taken during the darting. |
| Huunadjusted_stddev | HUMERUSES.Huunadjusted (computed) | The standard deviation of the unadjusted humerus length measurements related to the given Dartid -- the standard deviation of the unadjusted humerus length measurements taken during the darting. |
Contains one row for every unique Dartid value in the PCVS table.[274] Each row statistically summarizes the PCVS rows having the common Dartid value.
This view is useful when joined with the DARTINGS table on Dartid.
Figure 6.120. Query Defining the PCV_STATS View
SELECT pcvs.dartid AS dartid
, count(*) AS pcvsamps
, avg(pcvs.pcv) AS pcv_mean
, stddev(pcvs.pcv) AS pcv_stddev
FROM pcvs
GROUP BY pcvs.dartid;
Table 6.57. Columns in the PCV_STATS View
| Column | From | Description |
|---|---|---|
| Dartid | PCVS.Dartid | Identifier of the darting event. |
| PCVsamps | Computed | Number of PCVS rows having the given Dartid value -- the number of PCV measurements taken during the darting. |
| PCV_mean | PCVS.PCV (computed) | The arithmetic mean of the PCV measurements related to the given Dartid -- the mean of the PCV measurements taken during the darting. |
| PCV_stddev | PCVS.PCV (computed) | The standard deviation of the PCV measurements related to the given Dartid -- the standard deviation of the PCV measurements taken during the darting. |
Contains one row for every unique Dartid value in the TESTES_ARC table.[275] Each row statistically summarizes the TESTES_ARC rows having the common Dartid value.
This view is useful when joined with the DARTINGS table on Dartid.
Figure 6.122. Query Defining the TESTES_ARC_STATS View
SELECT testesdartids.dartid AS dartid
, testesllength.testllengthsamps AS testllengthsamps
, testesllength.testllength_mean AS testllength_mean
, testesllength.testllength_stddev AS testllength_stddev
, testeslwidth.testlwidthsamps AS testlwidthsamps
, testeslwidth.testlwidth_mean AS testlwidth_mean
, testeslwidth.testlwidth_stddev AS testlwidth_stddev
, testesrlength.testrlengthsamps AS testrlengthsamps
, testesrlength.testrlength_mean AS testrlength_mean
, testesrlength.testrlength_stddev AS testrlength_stddev
, testesrwidth.testrwidthsamps AS testrwidthsamps
, testesrwidth.testrwidth_mean AS testrwidth_mean
, testesrwidth.testrwidth_stddev AS testrwidth_stddev
FROM (SELECT testes_arc.dartid
FROM testes_arc
GROUP BY testes_arc.dartid)
AS testesdartids
LEFT OUTER JOIN
(SELECT testes_arc.dartid AS llengthdartid
, count(*) AS testllengthsamps
, avg(testes_arc.testlength) AS testllength_mean
, stddev(testes_arc.testlength) AS testllength_stddev
FROM testes_arc
WHERE testes_arc.testside = 'L'
AND testes_arc.testlength IS NOT NULL
GROUP BY testes_arc.dartid)
AS testesllength
ON testesllength.llengthdartid = testesdartids.dartid
LEFT OUTER JOIN
(SELECT testes_arc.dartid AS lwidthdartid
, count(*) AS testlwidthsamps
, avg(testes_arc.testwidth) AS testlwidth_mean
, stddev(testes_arc.testwidth) AS testlwidth_stddev
FROM testes_arc
WHERE testes_arc.testside = 'L'
AND testes_arc.testwidth IS NOT NULL
GROUP BY testes_arc.dartid)
AS testeslwidth
ON testeslwidth.lwidthdartid = testesdartids.dartid
LEFT OUTER JOIN
(SELECT testes_arc.dartid AS rlengthdartid
, count(*) AS testrlengthsamps
, avg(testes_arc.testlength) AS testrlength_mean
, stddev(testes_arc.testlength) AS testrlength_stddev
FROM testes_arc
WHERE testes_arc.testside = 'R'
AND testes_arc.testlength IS NOT NULL
GROUP BY testes_arc.dartid)
AS testesrlength
ON testesrlength.rlengthdartid = testesdartids.dartid
LEFT OUTER JOIN
(SELECT testes_arc.dartid AS rwidthdartid
, count(*) AS testrwidthsamps
, avg(testes_arc.testwidth) AS testrwidth_mean
, stddev(testes_arc.testwidth) AS testrwidth_stddev
FROM testes_arc
WHERE testes_arc.testside = 'R'
AND testes_arc.testwidth IS NOT NULL
GROUP BY testes_arc.dartid)
AS testesrwidth
ON testesrwidth.rwidthdartid = testesdartids.dartid;
Table 6.58. Columns in the TESTES_ARC_STATS View
| Column | From | Description |
|---|---|---|
| Dartid | TESTES_ARC.Dartid (computed) | Identifier of the darting event. |
| Testllengthsamps | Computed | Number of TESTES_ARC rows having the
given Dartid value and also having a TESTES_ARC.Testside value
of L and a non-NULL
TESTES_ARC.Testlength
value -- the number of left testicle length
measurements taken during the darting.[a] |
| Testllength_mean | TESTES_ARC.Testlength (computed) | The arithmetic mean of the left testicle length measurements related to the given Dartid -- the mean of the left testicle length measurements taken during the darting. |
| Testllength_stddev | TESTES_ARC.Testlength (computed) | The standard deviation of the left testicle length measurements related to the given Dartid -- the standard deviation of the left testicle length measurements taken during the darting. |
| Testlwidthsamps | Computed | Number of TESTES_ARC rows having the
given Dartid value and also having a TESTES_ARC.Testside value
of L and a non-NULL
TESTES_ARC.Testwidth value
-- the number of left testicle width measurements
taken during the darting.[b] |
| Testlwidth_mean | TESTES_ARC.Testwidth (computed) | The arithmetic mean of the left testicle width measurements related to the given Dartid -- the mean of the left testicle width measurements taken during the darting. |
| Testlwidth_stddev | TESTES_ARC.Testwidth (computed) | The standard deviation of the left testicle width measurements related to the given Dartid -- the standard deviation of the left testicle width measurements taken during the darting. |
| Testrlengthsamps | Computed | Number of TESTES_ARC rows having the
given Dartid value and also having a TESTES_ARC.Testside value
of R and a
non-NULL TESTES_ARC.Testlength
value -- the number of right testicle length
measurements taken during the darting.[c] |
| Testrlength_mean | TESTES_ARC.Testlength (computed) | The arithmetic mean of the right testicle length measurements related to the given Dartid -- the mean of the right testicle length measurements taken during the darting. |
| Testrlength_stddev | TESTES_ARC.Testlength (computed) | The standard deviation of the right testicle length measurements related to the given Dartid -- the standard deviation of the right testicle length measurements taken during the darting. |
| Testrwidthsamps | Computed | Number of TESTES_ARC rows having the
given Dartid value and also having a TESTES_ARC.Testside value
of R and a
non-NULL TESTES_ARC.Testwidth value
-- the number of right testicle width measurements
taken during the darting.[d] |
| Testrwidth_mean | TESTES_ARC.Testwidth (computed) | The arithmetic mean of the right testicle width measurements related to the given Dartid -- the mean of the right testicle width measurements taken during the darting. |
| Testrwidth_stddev | TESTES_ARC.Testwidth (computed) | The standard deviation of the right testicle width measurements related to the given Dartid -- the standard deviation of the right testicle width measurements taken during the darting. |
[a] [b] [c] [d] | ||
Contains one row for every unique Dartid value in the TESTES_DIAM table.[276] Each row statistically summarizes the TESTES_DIAM rows having the common Dartid value.
This view is useful when joined with the DARTINGS table on Dartid.
Figure 6.124. Query Defining the TESTES_DIAM_STATS View
SELECT testesdartids.dartid AS dartid
, testesllength.testllengthsamps AS testllengthsamps
, testesllength.testllength_mean AS testllength_mean
, testesllength.testllength_stddev AS testllength_stddev
, testeslwidth.testlwidthsamps AS testlwidthsamps
, testeslwidth.testlwidth_mean AS testlwidth_mean
, testeslwidth.testlwidth_stddev AS testlwidth_stddev
, testesrlength.testrlengthsamps AS testrlengthsamps
, testesrlength.testrlength_mean AS testrlength_mean
, testesrlength.testrlength_stddev AS testrlength_stddev
, testesrwidth.testrwidthsamps AS testrwidthsamps
, testesrwidth.testrwidth_mean AS testrwidth_mean
, testesrwidth.testrwidth_stddev AS testrwidth_stddev
FROM (SELECT testes_diam.dartid
FROM testes_diam
GROUP BY testes_diam.dartid)
AS testesdartids
LEFT OUTER JOIN
(SELECT testes_diam.dartid AS llengthdartid
, count(*) AS testllengthsamps
, avg(testes_diam.testlength) AS testllength_mean
, stddev(testes_diam.testlength) AS testllength_stddev
FROM testes_diam
WHERE testes_diam.testside = 'L'
AND testes_diam.testlength IS NOT NULL
GROUP BY testes_diam.dartid)
AS testesllength
ON testesllength.llengthdartid = testesdartids.dartid
LEFT OUTER JOIN
(SELECT testes_diam.dartid AS lwidthdartid
, count(*) AS testlwidthsamps
, avg(testes_diam.testwidth) AS testlwidth_mean
, stddev(testes_diam.testwidth) AS testlwidth_stddev
FROM testes_diam
WHERE testes_diam.testside = 'L'
AND testes_diam.testwidth IS NOT NULL
GROUP BY testes_diam.dartid)
AS testeslwidth
ON testeslwidth.lwidthdartid = testesdartids.dartid
LEFT OUTER JOIN
(SELECT testes_diam.dartid AS rlengthdartid
, count(*) AS testrlengthsamps
, avg(testes_diam.testlength) AS testrlength_mean
, stddev(testes_diam.testlength) AS testrlength_stddev
FROM testes_diam
WHERE testes_diam.testside = 'R'
AND testes_diam.testlength IS NOT NULL
GROUP BY testes_diam.dartid)
AS testesrlength
ON testesrlength.rlengthdartid = testesdartids.dartid
LEFT OUTER JOIN
(SELECT testes_diam.dartid AS rwidthdartid
, count(*) AS testrwidthsamps
, avg(testes_diam.testwidth) AS testrwidth_mean
, stddev(testes_diam.testwidth) AS testrwidth_stddev
FROM testes_diam
WHERE testes_diam.testside = 'R'
AND testes_diam.testwidth IS NOT NULL
GROUP BY testes_diam.dartid)
AS testesrwidth
ON testesrwidth.rwidthdartid = testesdartids.dartid;
Table 6.59. Columns in the TESTES_DIAM_STATS View
| Column | From | Description |
|---|---|---|
| Dartid | TESTES_DIAM.Dartid (computed) | Identifier of the darting event. |
| Testllengthsamps | Computed | Number of TESTES_DIAM rows having the
given Dartid value and also having a TESTES_DIAM.Testside value
of L and a non-NULL
TESTES_DIAM.Testlength
value -- the number of left testicle length
measurements taken during the darting.[a] |
| Testllength_mean | TESTES_DIAM.Testlength (computed) | The arithmetic mean of the left testicle length measurements related to the given Dartid -- the mean of the left testicle length measurements taken during the darting. |
| Testllength_stddev | TESTES_DIAM.Testlength (computed) | The standard deviation of the left testicle length measurements related to the given Dartid -- the standard deviation of the left testicle length measurements taken during the darting. |
| Testlwidthsamps | Computed | Number of TESTES_DIAM rows having the
given Dartid value and also having a TESTES_DIAM.Testside value
of L and a non-NULL
TESTES_DIAM.Testwidth value
-- the number of left testicle width measurements
taken during the darting.[b] |
| Testlwidth_mean | TESTES_DIAM.Testwidth (computed) | The arithmetic mean of the left testicle width measurements related to the given Dartid -- the mean of the left testicle width measurements taken during the darting. |
| Testlwidth_stddev | TESTES_DIAM.Testwidth (computed) | The standard deviation of the left testicle width measurements related to the given Dartid -- the standard deviation of the left testicle width measurements taken during the darting. |
| Testrlengthsamps | Computed | Number of TESTES_DIAM rows having the
given Dartid value and also having a TESTES_DIAM.Testside value
of R and a
non-NULL TESTES_DIAM.Testlength
value -- the number of right testicle length
measurements taken during the darting.[c] |
| Testrlength_mean | TESTES_DIAM.Testlength (computed) | The arithmetic mean of the right testicle length measurements related to the given Dartid -- the mean of the right testicle length measurements taken during the darting. |
| Testrlength_stddev | TESTES_DIAM.Testlength (computed) | The standard deviation of the right testicle length measurements related to the given Dartid -- the standard deviation of the right testicle length measurements taken during the darting. |
| Testrwidthsamps | Computed | Number of TESTES_DIAM rows having the
given Dartid value and also having a TESTES_DIAM.Testside value
of R and a
non-NULL TESTES_DIAM.Testwidth value
-- the number of right testicle width measurements
taken during the darting.[d] |
| Testrwidth_mean | TESTES_DIAM.Testwidth (computed) | The arithmetic mean of the right testicle width measurements related to the given Dartid -- the mean of the right testicle width measurements taken during the darting. |
| Testrwidth_stddev | TESTES_DIAM.Testwidth (computed) | The standard deviation of the right testicle width measurements related to the given Dartid -- the standard deviation of the right testicle width measurements taken during the darting. |
[a] [b] [c] [d] | ||
Contains one row for every unique Dartid value in the ULNAS table.[277] Each row statistically summarizes the ULNAS rows having the common Dartid value.
This view is useful when joined with the DARTINGS table on Dartid.
Figure 6.126. Query Defining the ULNA_STATS View
SELECT ulnas.dartid AS dartid
, count(*) AS ulsamps
, avg(ulnas.ullength) AS ullength_mean
, stddev(ulnas.ullength) AS ullength_stddev
, avg(ulnas.ulunadjusted) AS ulunadjusted_mean
, stddev(ulnas.ulunadjusted) AS ulunadjusted_stddev
FROM ulnas
GROUP BY ulnas.dartid;
Table 6.60. Columns in the ULNA_STATS View
| Column | From | Description |
|---|---|---|
| Dartid | ULNAS.Dartid | Identifier of the darting event. |
| Ulsamps | Computed | Number of ULNAS rows having the given Dartid value -- the number of ulna length measurements taken during the darting. |
| Ullength_mean | ULNAS.Ullength (computed) | The arithmetic mean of the ulna length measurements related to the given Dartid -- the mean of the ulna length measurements taken during the darting. |
| Ullength_stddev | ULNAS.Ullength (computed) | The standard deviation of the ulna length measurements related to the given Dartid -- the standard deviation of the ulna length measurements taken during the darting. |
| Ulunadjusted_mean | ULNAS.Ulunadjusted (computed) | The arithmetic mean of the unadjusted ulna length measurements related to the given Dartid -- the mean of the unadjusted ulna length measurements taken during the darting. |
| Ulunadjusted_stddev | ULNAS.Ulunadjusted (computed) | The standard deviation of the unadjusted ulna length measurements related to the given Dartid -- the standard deviation of the unadjusted ulna length measurements taken during the darting. |
Contains one row for every unique Dartid value in the VAGINAL_PHS table.[278] Each row statistically summarizes the VAGINAL_PHS rows having the common Dartid value.
This view is useful when joined with the DARTINGS table on Dartid.
Figure 6.128. Query Defining the VAGINAL_PH_STATS View
SELECT vaginal_phs.dartid AS dartid
, count(*) AS vpsamps
, avg(vaginal_phs.ph) AS vp_mean
, stddev(vaginal_phs.ph) AS vp_stddev
FROM vaginal_phs
GROUP BY vaginal_phs.dartid;
Table 6.61. Columns in the VAGINAL_PH_STATS View
| Column | From | Description |
|---|---|---|
| Dartid | VAGINAL_PHS.Dartid | Identifier of the darting event. |
| VPsamps | Computed | Number of VAGINAL_PHS rows having the given Dartid value — the number of vaginal pH measurements taken during the darting. |
| VP_mean | VAGINAL_PHS.PH (computed) | The arithmetic mean of the vaginal pH measurements related to the given Dartid — the mean of the vaginal pH measurements taken during the darting. |
| VP_stddev | VAGINAL_PHS.PH (computed) | The standard deviation of the vaginal pH measurements related to the given Dartid — the standard deviation of the vaginal pH measurements taken during the darting. |
Contains one row for every LOCATIONS row whose Location is not used in NUCACID_DATA or TISSUE_DATA. That is, it contains one row for every location that is not occupied by a nucleic acid or tissue sample.
Tip
Use this view when looking for locations available to store new samples.
Caution
This view makes no attempt to treat non-unique
locations (those whose Is_Unique
is FALSE) differently from unique ones. A non-unique
location that might in reality be available will not appear
in this view if it is already in use in NUCACID_DATA or TISSUE_DATA.
Figure 6.130. Query Defining the LOCATIONS_FREE View
SELECT locations.locid AS locid
, locations.institution AS institution
, locations.location AS location
, locations.is_unique AS is_unique
FROM locations
WHERE NOT EXISTS (SELECT 1
FROM tissue_data
WHERE tissue_data.locid = locations.locid)
AND NOT EXISTS (SELECT 1
FROM nucacid_data
WHERE nucacid_data.locid = locations.locid);
Table 6.62. Columns in the LOCATIONS_FREE View
| Column | From | Description |
|---|---|---|
| LocId | LOCATIONS.LocId | Identifier for the row |
| Institution | LOCATIONS.Institution | Organization, building, etc. describing the locale of this row's Location |
| Location | LOCATIONS.Location | Specific place/position available for a sample. |
| Is_Unique | LOCATIONS.Is_Unique | Whether or not this location can be used more than once. |
Contains one row for every NUCACID_CONC_DATA row. This view shows all the data from NUCACID_CONC_DATA, but also includes a descriptive column from NUCACID_CONC_METHODS to clarify the meaning of the Conc_Method column.
This view is also useful for adding data. New quantifications can be inserted using a sample's local ID's instead of their NAId.
Tip
Use this view instead of the NUCACID_CONC_DATA table.
Figure 6.132. Query Defining the NUCACID_CONCS View
SELECT nucacid_conc_data.nacid AS nacid
, nucacid_conc_data.naid AS naid
, local_1.localid AS localid_1
, local_2.localid AS localid_2
, nucacid_conc_data.conc_method AS conc_method
, nucacid_conc_methods.descr AS method_descr
, nucacid_conc_data.conc_date AS conc_date
, nucacid_conc_data.quantity AS quantity
, nucacid_conc_data.unit AS unit
FROM nucacid_conc_data
JOIN nucacid_conc_methods
ON nucacid_conc_methods.conc_method = nucacid_conc_data.conc_method
LEFT JOIN nucacid_local_ids AS local_1
ON local_1.naid = nucacid_conc_data.naid
AND local_1.institution = 1
LEFT JOIN nucacid_local_ids AS local_2
ON local_2.naid = nucacid_conc_data.naid
AND local_2.institution = 2;
Table 6.63. Columns in the NUCACID_CONCS View
| Column | From | Description |
|---|---|---|
| NACId | NUCACID_CONC_DATA.NACId | The unique identifier for this quantification. |
| NAId | NUCACID_CONC_DATA.NAId | The unique identifier for the quantified sample. |
| LocalId_1 | NUCACID_LOCAL_IDS.LocalId | The local identifier used for this sample at Institution #1, if any. |
| LocalId_2 | NUCACID_LOCAL_IDS.LocalId | The local identifier used for this sample at Institution #2, if any. |
| Conc_Method | NUCACID_CONC_DATA.Conc_Method | The method of quantification used to determine this concentration. |
| Method_Descr | NUCACID_CONC_METHODS.Descr | A textual description of the quantification method used to determine this concentration. |
| Conc_Date | NUCACID_CONC_DATA.Conc_Date | The date of the quantification. |
| Quantity | NUCACID_CONC_DATA.Quantity | The first half of the concentration: the number. |
| Unit | NUCACID_CONC_DATA.Unit | The second half of the concentration: the unit. |
Inserting a row into NUCACID_CONCS inserts a row into NUCACID_CONC_DATA as expected.
If no NAId is provided, one or both LocalId columns can be provided instead to look up the intended NAId. If LocalId_1 and/or LocalId_2 values are provided, these must be related to a single NUCACID_LOCAL_IDS.NAId value. If a NAId value is also provided, it must equal that single NAId that is related to the provided LocalId column(s).
At least one of either the Conc_Method or Method_Descr columns must be provided to determine the correct value to insert into NUCACID_CONC_DATA.Conc_Method. If Method_Descr is provided, it is used to look up the appropriate Conc_Method value from NUCACID_CONC_METHODS. If both are provided, the provided values must be related in NUCACID_CONC_METHODS.
Updating a row in NUCACID_CONCS updates the underlying NUCACID_CONC_DATA row, as discussed below.
The NAId may be updated in this view via updates to the NAId column only. Updates to the LocalId_1 and LocalId_2 columns result in an error[279].
A row's underlying NUCACID_CONC_DATA.Conc_Method may be updated in this view via updates to the Conc_Method or Method_Descr columns. If more than one of these values is updated, all the newly-updated values must be related, as discussed above.
Updating Conc_Method, Conc_Date, Quantity, or Unit updates the columns in the underlying NUCACID_CONC_DATA row, as expected.
Deleting a row from NUCACID_CONCS deletes a row from NUCACID_CONC_DATA as expected.
Contains one row for every NUCACID_SOURCES row. This view includes several subqueries from NUCACID_LOCAL_IDS, so that each sample and source are shown with their "local" identifiers. This view can be used to upload data.
Tip
Use this view instead of the NUCACID_SOURCES table.
Figure 6.134. Query Defining the NUCACID_SOURCES_EXT View
SELECT nucacid_sources.naid AS naid
, na_1.localid AS na_1
, na_2.localid AS na_2
, nucacid_sources.source_naid AS source_naid
, src_1.localid AS src_1
, src_2.localid AS src_2
, nucacid_sources.relationship
FROM nucacid_sources
LEFT JOIN nucacid_local_ids AS na_1
ON na_1.naid = nucacid_sources.naid
AND na_1.institution = 1
LEFT JOIN nucacid_local_ids AS na_2
ON na_2.naid = nucacid_sources.naid
AND na_2.institution = 2
LEFT JOIN nucacid_local_ids AS src_1
ON src_1.naid = nucacid_sources.source_naid
AND src_1.institution = 1
LEFT JOIN nucacid_local_ids AS src_2
ON src_2.naid = nucacid_sources.source_naid
AND src_2.institution = 2;
Table 6.64. Columns in the NUCACID_SOURCES_EXT View
| Column | From | Description |
|---|---|---|
| NAId | NUCACID_SOURCES.NAId | Identifier for this sample. |
| NA_1 | NUCACID_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #1. |
| NA_2 | NUCACID_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #2. |
| Source_NAId | NUCACID_SOURCES.Source_NAId | Identifier for this source. |
| Src_1 | NUCACID_LOCAL_IDS.LocalId | The local identifier, if any, used for this source at Institution #1. |
| Src_2 | NUCACID_LOCAL_IDS.LocalId | The local identifier, if any, used for this source at Institution #2. |
| Relationship | NUCACID_SOURCES.Relationship | A textual description of how this sample is related to this source. |
Inserting a row into NUCACID_SOURCES_EXT inserts a row in NUCACID_SOURCES, as expected.
To identify the nucleic acid sample, only one of the NAId, NA_1, and NA_2 columns must be provided. If more than one is provided, they must match in NUCACID_LOCAL_IDS.
To identify the source, only one of the Source_NAId, Src_1, and Src_2 columns must be provided. If more than one is provided, they must match in NUCACID_LOCAL_IDS.
Updating a row in NUCACID_SOURCES_EXT updates the underlying row in NUCACID_SOURCES, as expected.
If updating the nucleic acid sample, update only one of the NAId, NA_1, or NA_2 columns. The underlying NUCACID_SOURCES.NAId will be updated as expected.
If updating the source, update only one of the Source_NAId, Src_1, or Src_2 columns. The underlying NUCACID_SOURCES.Source_NAId will be updated as expected.
Deleting a row in NUCACID_SOURCES_EXT deletes the underlying row in NUCACID_SOURCES, as expected.
Contains one row for every NUCACID_DATA row. This view includes columns from BIOGRAPH, LOCATIONS, NUCACID_CREATORS, NUCACID_LOCAL_IDS, NUCACID_SOURCES, TISSUE_DATA, and UNIQUE_INDIVS in order to portray information about nucleic acid samples in a more user-friendly format than that in NUCACID_DATA. This view can also be used to upload data.
Tip
Use this view — or the NUCACIDS_W_CONC view if the sample's concentration is important to you — instead of the NUCACID_DATA table.
When uploading data
with this view, it is an error if creator initials cannot be
unambiguously interpreted. In the admittedly-unlikely event
that there is a creator whose initials legitimately include
the separator character
"/", this creator's
initials cannot be inserted via this view. In this case, the
offending creator code must be removed from the data, then
manually inserted into NUCACID_CREATORS.
Figure 6.136. Query Defining the NUCACIDS View
WITH concat_creators AS (SELECT naid
, string_agg(creator, '/' ORDER BY naid, nacrid) AS created_by
FROM nucacid_creators
GROUP BY naid)
SELECT nucacid_data.naid AS naid
, nucacid_data.tid AS tid
, nucacid_data.locid AS locid
, locations.institution AS institution
, locations.location AS location
, local_1.localid AS localid_1
, local_2.localid AS localid_2
, nucacid_data.uiid AS uiid
, unique_indivs.popid AS popid
, unique_indivs.individ AS individ
, biograph.sname AS sname
, nucacid_data.name_on_tube AS name_on_tube
, nucacid_data.nucacid_type AS nucacid_type
, tissue_data.tissue_type AS tissue_type
, nucacid_data.creation_date AS creation_date
, concat_creators.created_by AS created_by
, nucacid_data.creation_method AS creation_method
, COUNT(nucacid_sources.*) AS na_sources
, nucacid_data.initial_vol_ul AS initial_vol_ul
, nucacid_data.actual_vol_ul AS actual_vol_ul
, nucacid_data.actual_vol_date AS actual_vol_date
, nucacid_data.multi_indivs AS multi_indivs
, nucacid_data.multi_tids AS multi_tids
, nucacid_data.notes AS notes
FROM nucacid_data
JOIN locations
ON locations.locid = nucacid_data.locid
LEFT JOIN tissue_data
ON tissue_data.tid = nucacid_data.tid
LEFT JOIN unique_indivs
ON unique_indivs.uiid = nucacid_data.uiid
LEFT JOIN biograph
ON biograph.bioid::text = unique_indivs.individ
AND unique_indivs.popid = 1
LEFT JOIN nucacid_local_ids AS local_1
ON local_1.naid = nucacid_data.naid
AND local_1.institution = 1
LEFT JOIN nucacid_local_ids AS local_2
ON local_2.naid = nucacid_data.naid
AND local_2.institution = 2
LEFT JOIN nucacid_sources
ON nucacid_sources.naid = nucacid_data.naid
LEFT JOIN concat_creators
ON concat_creators.naid = nucacid_data.naid
GROUP BY nucacid_data.naid
, nucacid_data.tid
, nucacid_data.locid
, locations.institution
, locations.location
, local_1.localid
, local_2.localid
, nucacid_data.uiid
, unique_indivs.popid
, unique_indivs.individ
, biograph.sname
, nucacid_data.name_on_tube
, nucacid_data.nucacid_type
, tissue_data.tissue_type
, nucacid_data.creation_date
, concat_creators.created_by
, nucacid_data.creation_method
, nucacid_data.initial_vol_ul
, nucacid_data.actual_vol_ul
, nucacid_data.actual_vol_date
, nucacid_data.multi_indivs
, nucacid_data.multi_tids
, nucacid_data.notes;
Table 6.65. Columns in the NUCACIDS View
| Column | From | Description |
|---|---|---|
| NAId | NUCACID_DATA.NAId | Identifier for this sample. |
| TId | NUCACID_DATA.TId | Identifier for this nucleic acid sample's source tissue sample, if there's only one. |
| LocId | NUCACID_DATA.LocId | Identifier for this sample's Institution-Location pair. |
| Institution | LOCATIONS.Institution | Identifier for this sample's locale. |
| Location | LOCATIONS.Location | The current place/position of the sample. |
| LocalId_1 | NUCACID_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #1. |
| LocalId_2 | NUCACID_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #2. |
| UIId | NUCACID_DATA.UIId | Identifier for the source individual, if recorded. |
| PopId | UNIQUE_INDIVS.PopId | Identifier for the population of the source individual, if the source individual is recorded. |
| IndivId | UNIQUE_INDIVS.IndivId | Name/ID of the source individual, if the source individual is recorded. |
| Sname | BIOGRAPH.Sname | Sname of the source individual, if the source individual is recorded and they have an Sname. |
| Name_on_Tube | NUCACID_DATA.Name_on_Tube | Name/identifier written on the sample's label. |
| NucAcid_Type | NUCACID_DATA.NucAcid_Type | The nucleic acid sample type. |
| Tissue_Type | TISSUE_DATA.Tissue_Type | The source tissue's sample type. |
| Creation_Date | NUCACID_DATA.Creation_Date | Date that the sample was created. |
| Created_By | NUCACID_CREATORS.Creator | Initials of all the personnel involved with
the creation of this sample concatenated into a
single string, ordered by their related NUCACID_CREATORS.NACrId and separated by
a "/". If no
related creators, then NULL. |
| Creation_Method | NUCACID_DATA.Creation_Method | The method used to create the sample. |
| NA_Sources | COUNT(NUCACID_SOURCES.*) | The number of rows in NUCACID_SOURCES with this NAId. |
| Initial_Vol_ul | NUCACID_DATA.Initial_Vol_ul | Volume in microliters of the sample when first created. |
| Actual_Vol_ul | NUCACID_DATA.Actual_Vol_ul | The amount of sample (in microliters) remaining in the tube, as of the Actual_Vol_Date. |
| Actual_Vol_Date | NUCACID_DATA.Actual_Vol_Date | The date that the Actual_Vol_ul was determined. |
| Multi_Indivs | NUCACID_DATA.Multi_Indivs | Boolean indicating whether or not this sample is attributed to more than one individual. |
| Multi_TIds | NUCACID_DATA.Multi_TIds | Boolean indicating whether or not this sample has more than one source tissue. |
| Notes | NUCACID_DATA.Notes | Miscellaneous notes about the sample. |
Inserting a row into NUCACIDS inserts a row into NUCACID_DATA. Additional rows may be inserted into NUCACID_CREATORS or NUCACID_LOCAL_IDS, as discussed below. This view does not insert or update rows in NUCACID_SOURCES.
For each
"/"-separated creator
provided in the Created_By column, one row is inserted
into the NUCACID_CREATORS table, with
the related NAId. A NULL
Created_By column is interpreted to mean that there are no
rows to add to NUCACID_CREATORS; it does
not result in a new NUCACID_CREATORS row with a NULL Creator value.
When either or
both of the LocalId_1 and LocalId_2 columns is not NULL,
a row is inserted into NUCACID_LOCAL_IDS
for each non-NULL value provided. The new NUCACID_LOCAL_IDS.LocalId is the provided
LocalId_N value, and the new Institution is
1 (for LocalId_1) or
2 (for
LocalId_2).
To indicate a sample's current locale and location, either the LocId column or both the Institution and Location columns must be provided. If all three are provided, the Institution and Location must be equal to the related columns in LOCATIONS for the provided LocId.
When the sample should be attributed to a single individual, that individual must be identified via the UIId, PopId, IndivId, and/or Sname columns. It is not necessary to provide values for all of those columns; there must only be enough information provided to identify a single UIId. Specifically: there must be a UIId, a PopId and an IndivId, or an Sname. When more than one of those is provided, all provided values must be related to the same UNIQUE_INDIVS.UIId, which is inserted as the new NUCACIDS.UIId.
When the sample should not be attributed to a single
individual — the NUCACIDS.UIId will be NULL — the
UIId, PopId, IndivId, and Sname must all be NULL.
It is not necessary to provide Tissue_Type. If provided, it must match the related TISSUE_DATA.Tissue_Type value.
The NA_SOURCES column is calculated automatically.
When inserting data, there cannot possibly be any rows in
NUCACID_SOURCES yet, so if a value is
provided for this column it must be
0.
Updating a row in NUCACIDS updates the underlying row in NUCACID_DATA, as expected. Related rows in NUCACID_CREATORS and NUCACID_LOCAL_IDS may be inserted, updated, or deleted, as discussed below. This view does not insert or update rows in NUCACID_SOURCES.
When an update changes the Created_By column, all prior rows in NUCACID_CREATORS are deleted, and new rows are inserted as described above. When an update doesn't change the Created_By column, the related data in NUCACID_CREATORS are unaffected.
When LocalId_1 or LocalId_2 is changed, the related
NUCACID_LOCAL_IDS.LocalId value is also
changed as expected, except when the "old" or "new" value
is NULL. If the change is from NULL to non-NULL, a
new NUCACID_LOCAL_IDS row is inserted,
as discussed above. If
from non-NULL to NULL, the related NUCACID_LOCAL_IDS row is deleted.
Updating the Institution and Location columns updates the related LocId column, as expected.
Updating a sample's UIId can be done by updating the UIId, PopId and IndivId, and/or Sname columns. Any such updates must correspond to exactly one UIId, as discussed above.
The NA_SOURCES column is calculated automatically and cannot be manually changed. When updating data, any value provided for this column is silently ignored.
Deleting a row from NUCACIDS deletes the underlying row from NUCACID_DATA, as expected. Related rows in NUCACID_CREATORS, NUCACID_LOCAL_IDS, and NUCACID_SOURCES, if any, are also deleted.
This view contains one row for every row in NUCACID_DATA. It includes columns from BIOGRAPH, LOCATIONS, NUCACID_CREATORS, NUCACID_LOCAL_IDS, NUCACID_SOURCES, TISSUE_DATA, and UNIQUE_INDIVS, as in the NUCACIDS view. It also includes several additional columns derived from NUCACID_CONC_DATA that indicate the sample's concentration according to various specific quantification methods.
Tip
Use this view — or just NUCACIDS if the sample's concentration is not important to you — instead of the NUCACID_DATA table.
Warning
A nucleic acid sample's concentration may be
quantified more than once with the same method, so
this view shows only the concentration from the most recent
NUCACID_CONC_DATA.Conc_Date for each
method. Because of this, concentrations whose related Conc_Date is NULL are not
included in this view.
Figure 6.138. Query Defining the NUCACIDS_W_CONC View
WITH last_quants AS (SELECT DISTINCT
naid
, conc_method
, last_value(quantity) OVER w AS lastquantity
, last_value(unit) OVER w AS lastunit
, last_value(conc_date) OVER w AS lastdate
FROM nucacid_conc_data
WHERE conc_date IS NOT NULL
WINDOW w AS (PARTITION BY naid, conc_method
ORDER BY conc_date
RANGE BETWEEN UNBOUNDED PRECEDING
AND UNBOUNDED FOLLOWING))
, concat_creators AS (SELECT naid
, string_agg(creator, '/' ORDER BY naid, nacrid) AS created_by
FROM nucacid_creators
GROUP BY naid)
SELECT nucacid_data.naid AS naid
, nucacid_data.tid AS tid
, nucacid_data.locid AS locid
, locations.institution AS institution
, locations.location AS location
, local_1.localid AS localid_1
, local_2.localid AS localid_2
, nucacid_data.uiid AS uiid
, unique_indivs.popid AS popid
, unique_indivs.individ AS individ
, biograph.sname AS sname
, nucacid_data.name_on_tube AS name_on_tube
, nucacid_data.nucacid_type AS nucacid_type
, tissue_data.tissue_type AS tissue_type
, nucacid_data.creation_date AS creation_date
, concat_creators.created_by AS created_by
, nucacid_data.creation_method AS creation_method
, COUNT(nucacid_sources.*) AS na_sources
, nucacid_data.initial_vol_ul AS initial_vol_ul
, nucacid_data.actual_vol_ul AS actual_vol_ul
, nucacid_data.actual_vol_date AS actual_vol_date
, nucacid_data.multi_indivs AS multi_indivs
, nucacid_data.multi_tids AS multi_tids
, nucacid_data.notes AS notes
, CASE
WHEN qpcr.lastquantity IS NOT NULL
THEN convert_conc(qpcr.lastquantity, qpcr.lastunit, 'bb_pgul_unit')
ELSE NULL
END AS qpcr_pg_ul
, qpcr.lastdate AS qpcr_lastdate
, CASE
WHEN nanodrop.lastquantity IS NOT NULL
THEN convert_conc(nanodrop.lastquantity, nanodrop.lastunit, 'bb_ngul_unit')
ELSE NULL
END AS nanodrop_ng_ul
, nanodrop.lastdate AS nanodrop_lastdate
, CASE
WHEN qubit.lastquantity IS NOT NULL
THEN convert_conc(qubit.lastquantity, qubit.lastunit, 'bb_ngul_unit')
ELSE NULL
END AS qubit_ng_ul
, qubit.lastdate AS qubit_lastdate
, CASE
WHEN bioanalyzer.lastquantity IS NOT NULL
THEN convert_conc(bioanalyzer.lastquantity, bioanalyzer.lastunit, 'bb_ngul_unit')
ELSE NULL
END AS bioanalyzer_ng_ul
, bioanalyzer.lastdate AS bioanalyzer_lastdate
, CASE
WHEN quantit.lastquantity IS NOT NULL
THEN convert_conc(quantit.lastquantity, quantit.lastunit, 'bb_ngul_unit')
ELSE NULL
END AS quantit_ng_ul
, quantit.lastdate AS quantit_lastdate
FROM nucacid_data
JOIN locations
ON locations.locid = nucacid_data.locid
LEFT JOIN tissue_data
ON tissue_data.tid = nucacid_data.tid
LEFT JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON biograph.bioid::text = unique_indivs.individ
AND unique_indivs.popid = 1
LEFT JOIN nucacid_local_ids AS local_1
ON local_1.naid = nucacid_data.naid
AND local_1.institution = 1
LEFT JOIN nucacid_local_ids AS local_2
ON local_2.naid = nucacid_data.naid
AND local_2.institution = 2
LEFT JOIN nucacid_sources
ON nucacid_sources.naid = nucacid_data.naid
LEFT JOIN concat_creators
ON concat_creators.naid = nucacid_data.naid
LEFT JOIN last_quants AS qpcr
ON qpcr.conc_method = 1
AND qpcr.naid = nucacid_data.naid
LEFT JOIN last_quants AS nanodrop
ON nanodrop.conc_method = 2
AND nanodrop.naid = nucacid_data.naid
LEFT JOIN last_quants AS qubit
ON qubit.conc_method = 3
AND qubit.naid = nucacid_data.naid
LEFT JOIN last_quants AS bioanalyzer
ON bioanalyzer.conc_method = 4
AND bioanalyzer.naid = nucacid_data.naid
LEFT JOIN last_quants AS quantit
ON quantit.conc_method = 5
AND quantit.naid = nucacid_data.naid
GROUP BY nucacid_data.naid
, nucacid_data.tid
, nucacid_data.locid
, locations.institution
, locations.location
, local_1.localid
, local_2.localid
, nucacid_data.uiid
, unique_indivs.popid
, unique_indivs.individ
, biograph.sname
, nucacid_data.name_on_tube
, nucacid_data.nucacid_type
, tissue_data.tissue_type
, nucacid_data.creation_date
, concat_creators.created_by
, nucacid_data.creation_method
, nucacid_data.initial_vol_ul
, nucacid_data.actual_vol_ul
, nucacid_data.actual_vol_date
, nucacid_data.multi_indivs
, nucacid_data.multi_tids
, nucacid_data.notes
, qpcr.lastquantity
, qpcr.lastunit
, qpcr.lastdate
, nanodrop.lastquantity
, nanodrop.lastunit
, nanodrop.lastdate
, qubit.lastquantity
, qubit.lastunit
, qubit.lastdate
, bioanalyzer.lastquantity
, bioanalyzer.lastunit
, bioanalyzer.lastdate
, quantit.lastquantity
, quantit.lastunit
, quantit.lastdate;
Table 6.66. Columns in the NUCACIDS_W_CONC View
| Column | From | Description |
|---|---|---|
| NAId | NUCACID_DATA.NAId | Identifier for this sample. |
| TId | NUCACID_DATA.TId | Identifier for this nucleic acid sample's source tissue sample, if there's only one. |
| LocId | NUCACID_DATA.LocId | Identifier for this sample's Institution-Location pair. |
| Institution | LOCATIONS.Institution | Identifier for this sample's locale. |
| Location | LOCATIONS.Location | The current place/position of the sample. |
| LocalId_1 | NUCACID_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #1. |
| LocalId_2 | NUCACID_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #2. |
| UIId | NUCACID_DATA.UIId | Identifier for the source individual, if recorded. |
| PopId | UNIQUE_INDIVS.PopId | Identifier for the population of the source individual, if the source individual is recorded. |
| IndivId | UNIQUE_INDIVS.IndivId | Name/ID of the source individual, if the source individual is recorded. |
| Sname | BIOGRAPH.Sname | Sname of the source individual, if the source individual is recorded and they have an Sname. |
| Name_on_Tube | NUCACID_DATA.Name_on_Tube | Name/identifier written on the sample's label. |
| NucAcid_Type | NUCACID_DATA.NucAcid_Type | The nucleic acid sample type. |
| Tissue_Type | TISSUE_DATA.Tissue_Type | The source tissue's sample type. |
| Creation_Date | NUCACID_DATA.Creation_Date | Date that the sample was created. |
| Created_By | NUCACID_CREATORS.Creator | Initials of all the personnel involved with
the creation of this sample concatenated into a
single string, ordered by their related NUCACID_CREATORS.NACrId and separated by
a "/". If no
related creators, then NULL. |
| Creation_Method | NUCACID_DATA.Creation_Method | The method used to create the sample. |
| NA_Sources | COUNT(NUCACID_SOURCES.*) | The number of rows in NUCACID_SOURCES with this NAId. |
| Initial_Vol_ul | NUCACID_DATA.Initial_Vol_ul | Volume in microliters of the sample when first created. |
| Actual_Vol_ul | NUCACID_DATA.Actual_Vol_ul | The amount of sample (in microliters) remaining in the tube, as of the Actual_Vol_Date. |
| Actual_Vol_Date | NUCACID_DATA.Actual_Vol_Date | The date that the Actual_Vol_ul was determined. |
| Multi_Indivs | NUCACID_DATA.Multi_Indivs | Boolean indicating whether or not this sample is attributed to more than one individual. |
| Multi_TIds | NUCACID_DATA.Multi_TIds | Boolean indicating whether or not this sample has more than one source tissue. |
| Notes | NUCACID_DATA.Notes | Miscellaneous notes about the sample. |
| QPCR_Pg_ul | convert_conc(NUCACID_CONC_DATA.Quantity, NUCACID_CONC_DATA.Unit, 'PG/UL') | The concentration of this sample in pg/μL, according to the most recent quantitative PCR. |
| QPCR_LastDate | NUCACID_CONC_DATA.Conc_Date | The date of this row's QPCR_Pg_ul was determined; the date of the most recent QPCR. |
| Nanodrop_Ng_ul | convert_conc(NUCACID_CONC_DATA.Quantity, NUCACID_CONC_DATA.Unit, 'NG/UL') | The concentration of this sample in ng/μL, according to the most recent Nanodrop measurement. |
| Nanodrop_LastDate | NUCACID_CONC_DATA.Conc_Date | The date that this row's Nanodrop_Ng_ul was determined; the date of the most recent Nanodrop measurement. |
| Qubit_Ng_ul | convert_conc(NUCACID_CONC_DATA.Quantity, NUCACID_CONC_DATA.Unit, 'NG/UL') | The concentration of this sample in ng/μL, according to the most recent Qubit measurement. |
| Qubit_LastDate | NUCACID_CONC_DATA.Conc_Date | The date that this row's Qubit_Ng_ul was determined; the date of the most recent Qubit measurement. |
| Bioanalyzer_Ng_ul | convert_conc(NUCACID_CONC_DATA.Quantity, NUCACID_CONC_DATA.Unit, 'NG/UL') | The concentration of this sample in ng/μL, according to the most recent Bioanalyzer run. |
| Bioanalyzer_LastDate | NUCACID_CONC_DATA.Conc_Date | The date that this row's Bioanalyzer_Ng_ul was determined; the date of the most recent Bioanalyzer run. |
| Quantit_Ng_ul | convert_conc(NUCACID_CONC_DATA.Quantity, NUCACID_CONC_DATA.Unit, 'NG/UL') | The concentration of this sample in ng/μ/L, according to the most recent Quant-iT assay. |
| Quantit_LastDate | NUCACID_CONC_DATA.Conc_Date | The date that this row's Quantit_Ng_ul was determined; the date of the most-recent Quant-iT assay. |
Contains one row for every TISSUE_SOURCES row. This view includes several subqueries from TISSUE_LOCAL_IDS, so that each sample and source are shown with their "local" identifiers. This view can be used to upload data.
Tip
Use this view instead of the TISSUE_SOURCES table.
Figure 6.140. Query Defining the TISSUE_SOURCES_EXT View
SELECT tissue_sources.tid AS tid
, t_1.localid AS t_1
, t_2.localid AS t_2
, tissue_sources.source_tid AS source_tid
, src_1.localid AS src_1
, src_2.localid AS src_2
, tissue_sources.relationship
FROM tissue_sources
LEFT JOIN tissue_local_ids AS t_1
ON t_1.tid = tissue_sources.tid
AND t_1.institution = 1
LEFT JOIN tissue_local_ids AS t_2
ON t_2.tid = tissue_sources.tid
AND t_2.institution = 2
LEFT JOIN tissue_local_ids AS src_1
ON src_1.tid = tissue_sources.source_tid
AND src_1.institution = 1
LEFT JOIN tissue_local_ids AS src_2
ON src_2.tid = tissue_sources.source_tid
AND src_2.institution = 2;
Table 6.67. Columns in the TISSUE_SOURCES_EXT View
| Column | From | Description |
|---|---|---|
| TId | TISSUE_SOURCES.TId | Identifier for this sample. |
| T_1 | TISSUE_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #1. |
| T_2 | TISSUE_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #2. |
| Source_TId | TISSUE_SOURCES.Source_TId | Identifier for this source. |
| Src_1 | TISSUE_LOCAL_IDS.LocalId | The local identifier, if any, used for this source at Institution #1. |
| Src_2 | TISSUE_LOCAL_IDS.LocalId | The local identifier, if any, used for this source at Institution #2. |
| Relationship | TISSUE_SOURCES.Relationship | A textual description of how this sample is related to this source. |
Inserting a row into TISSUE_SOURCES_EXT inserts a row in TISSUE_SOURCES, as expected.
To identify the tissue sample, only one of the TId, T_1, and T_2 columns must be provided. If more than one is provided, they must match in TISSUE_LOCAL_IDS.
To identify the source, only one of the Source_TId, Src_1, and Src_2 columns must be provided. If more than one is provided, they must match in TISSUE_LOCAL_IDS.
Updating a row in TISSUE_SOURCES_EXT updates the underlying row in TISSUE_SOURCES, as expected.
If updating the tissue sample, update only one of the TId, T_1, or T_2 columns. The underlying TISSUE_SOURCES.TId will be updated as expected.
If updating the source, update only one of the Source_TId, Src_1, or Src_2 columns. The underlying TISSUE_SOURCES.Source_TId will be updated as expected.
Deleting a row in TISSUE_SOURCES_EXT deletes the underlying row in TISSUE_SOURCES, as expected.
Contains one row for every TISSUE_DATA row. This view includes columns from BIOGRAPH, LOCATIONS, TISSUE_LOCAL_IDS, and UNIQUE_INDIVS in order to portray information about tissue samples in a more user-friendly format than that in TISSUE_DATA. This view can also be used to upload data.
Tip
Use this view instead of the TISSUE_DATA table.
Figure 6.142. Query Defining the TISSUES View
SELECT tissue_data.tid AS tid
, tissue_data.locid AS locid
, locations.institution AS institution
, locations.location AS location
, local_1.localid AS localid_1
, local_2.localid AS localid_2
, tissue_data.uiid AS uiid
, unique_indivs.popid AS popid
, unique_indivs.individ AS individ
, biograph.sname AS sname
, tissue_data.name_on_tube AS name_on_tube
, tissue_data.collection_date AS collection_date
, tissue_data.collection_time AS collection_time
, tissue_data.tissue_type AS tissue_type
, tissue_data.storage_medium AS storage_medium
, tissue_data.misid_status AS misid_status
, tissue_data.collection_date_status AS collection_date_status
, tissue_data.multi_indivs AS multi_indivs
, COUNT(tissue_sources.*) AS tissue_sources
, tissue_data.notes AS notes
FROM tissue_data
JOIN locations
ON locations.locid = tissue_data.locid
LEFT JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON biograph.bioid::text = unique_indivs.individ
AND unique_indivs.popid = 1
LEFT JOIN tissue_local_ids AS local_1
ON local_1.tid = tissue_data.tid
AND local_1.institution = 1
LEFT JOIN tissue_local_ids AS local_2
ON local_2.tid = tissue_data.tid
AND local_2.institution = 2
LEFT JOIN tissue_sources
ON tissue_sources.tid = tissue_data.tid
GROUP BY tissue_data.tid
, tissue_data.locid
, locations.institution
, locations.location
, local_1.localid
, local_2.localid
, tissue_data.uiid
, unique_indivs.popid
, unique_indivs.individ
, biograph.sname
, tissue_data.name_on_tube
, tissue_data.collection_date
, tissue_data.collection_time
, tissue_data.tissue_type
, tissue_data.storage_medium
, tissue_data.misid_status
, tissue_data.collection_date_status
, tissue_data.multi_indivs
, tissue_data.notes;
Table 6.68. Columns in the TISSUES View
| Column | From | Description |
|---|---|---|
| TId | TISSUE_DATA.TId | Identifier for this sample. |
| LocId | TISSUE_DATA.LocId | Identifier for this sample's Institution-Location pair. |
| Institution | LOCATIONS.Institution | Identifier for this sample's locale. |
| Location | LOCATIONS.Location | The current place/position of the sample. |
| LocalId_1 | TISSUE_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #1. |
| LocalId_2 | TISSUE_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #2. |
| UIId | TISSUE_DATA.UIId | Identifier for the source individual, if recorded. |
| PopId | UNIQUE_INDIVS.PopId | Identifier for the population of the source individual, if the source individual is recorded. |
| IndivId | UNIQUE_INDIVS.IndivId | Name/ID of the source individual, if the source individual is recorded. |
| Sname | BIOGRAPH.Sname | Sname of the source individual, if the source individual is recorded and they have an Sname. |
| Name_on_Tube | TISSUE_DATA.Name_on_Tube | Name or ID of the source individual, according to the label on the tube. |
| Collection_Date | TISSUE_DATA.Collection_Date | Date that the sample was collected. |
| Collection_Time | TISSUE_DATA.Collection_Time | Time that the sample was collected. |
| Tissue_Type | TISSUE_DATA.Tissue_Type | The tissue sample type. |
| Storage_Medium | TISSUE_DATA.Storage_Medium | The medium used for storing the sample. |
| Misid_Status | TISSUE_DATA.Misid_Status | The mis-identification status of the sample, if the source individual is recorded. |
| Collection_Date_Status | TISSUE_DATA.Collection_Date_Status | The status of this Collection_Date |
| Multi_Indivs | TISSUE_DATA.Multi_Indivs | Boolean indicating whether this sample has tissue sources from more than one individual. |
| Tissue_Sources | COUNT(TISSUE_SOURCES.*) | The number of rows in TISSUE_SOURCES with this TId. |
| Notes | TISSUE_DATA.Notes | Miscellaneous notes about the sample. |
Inserting a row into TISSUES inserts a row into TISSUE_DATA. Additional rows may be inserted into TISSUE_LOCAL_IDS, as discussed below.
When either or both
of the LocalId_1 and LocalId_2 columns is not NULL, a
row is inserted into TISSUE_LOCAL_IDS
for each non-NULL value provided. The new TISSUE_LOCAL_IDS.LocalId is the provided
LocalId_N value, and the new Institution is
1 (for LocalId_1) or
2 (for
LocalId_2).
It is not necessary to provide all of the UIId, PopId, IndivId, and Sname columns; there must only be enough information provided to identify a single UIId. Specifically: there must be a UIId, a PopId and an IndivId, or an Sname. When more than one of those is provided, all provided values must be related to the same UNIQUE_INDIVS.UIId.
To indicate a sample's current locale and location, either the LocId column or both the Institution and Location columns must be provided. If all three are provided, the Institution and Location must be equal to the related columns in LOCATIONS for the provided LocId.
The Tissue_Sources column is calculated
automatically. When inserting data, there cannot possibly
be any rows in TISSUE_SOURCES yet, so if
a value is provided for this column it must be
0.
Updating a row in TISSUES updates the underlying row in TISSUE_DATA, as expected. Related rows in TISSUE_LOCAL_IDS may be inserted, updated, or deleted, as discussed below.
When LocalId_1 or LocalId_2 is changed, the related
TISSUE_LOCAL_IDS.LocalId value is also
changed, as expected. If this change is from NULL to
non-NULL, a new TISSUE_LOCAL_IDS row
is inserted, as discussed above. If
from non-NULL to NULL, the related TISSUE_LOCAL_IDS row is deleted.
Updating a sample's UIId can be done by updating the UIId, PopId and IndivId, and/or Sname columns. Any such updates must correspond to exactly one UIId, as discussed above.
Updating the Institution and Location columns updates the related LocId column, as expected.
The TISSUE_SOURCES column is calculated automatically and cannot be manually changed. When updating data, any value provided for this column is silently ignored.
Deleting a row from TISSUES deletes the underlying row from TISSUE_DATA, as expected. Related rows in TISSUE_LOCAL_IDS and TISSUE_SOURCES, if any, are also deleted.
Contains one row for every TISSUE_DATA row. This view includes columns from BIOGRAPH, LOCATIONS, TISSUE_LOCAL_IDS, UNIQUE_INDIVS, and HORMONE_SAMPLE_DATA in order to portray information about tissue samples in a more user-friendly format than that in TISSUE_DATA, especially samples that are used in hormone analysis. This view is also useful for uploading new tissue samples that will be used for hormone analysis; it provides a way to upload samples into TISSUE_DATA and HORMONE_SAMPLE_DATA simultaneously.
Figure 6.144. Query Defining the TISSUES_HORMONES View
SELECT tissue_data.tid AS tid
, tissue_data.locid AS locid
, locations.institution AS institution
, locations.location AS location
, local_1.localid AS localid_1
, local_2.localid AS localid_2
, tissue_data.uiid AS uiid
, unique_indivs.popid AS popid
, unique_indivs.individ AS individ
, biograph.sname AS sname
, tissue_data.name_on_tube AS name_on_tube
, tissue_data.collection_date AS collection_date
, tissue_data.collection_time AS collection_time
, tissue_data.tissue_type AS tissue_type
, tissue_data.storage_medium AS storage_medium
, tissue_data.misid_status AS misid_status
, tissue_data.collection_date_status AS collection_date_status
, tissue_data.multi_indivs AS multi_indivs
, COUNT(tissue_sources.*) AS tissue_sources
, tissue_data.notes AS notes
, hormone_sample_data.hsid AS hsid
, hormone_sample_data.fzdried_date AS fzdried_date
, hormone_sample_data.sifted_date AS sifted_date
, hormone_sample_data.avail_mass_g AS avail_mass_g
, hormone_sample_data.avail_date AS avail_date
, hormone_sample_data.comments AS comments
FROM tissue_data
JOIN locations
ON locations.locid = tissue_data.locid
LEFT JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON biograph.bioid::text = unique_indivs.individ
AND unique_indivs.popid = 1
LEFT JOIN tissue_local_ids AS local_1
ON local_1.tid = tissue_data.tid
AND local_1.institution = 1
LEFT JOIN tissue_local_ids AS local_2
ON local_2.tid = tissue_data.tid
AND local_2.institution = 2
LEFT JOIN tissue_sources
ON tissue_sources.tid = tissue_data.tid
LEFT JOIN hormone_sample_data
ON hormone_sample_data.tid = tissue_data.tid
GROUP BY tissue_data.tid
, tissue_data.locid
, locations.institution
, locations.location
, local_1.localid
, local_2.localid
, tissue_data.uiid
, unique_indivs.popid
, unique_indivs.individ
, biograph.sname
, tissue_data.name_on_tube
, tissue_data.collection_date
, tissue_data.collection_time
, tissue_data.tissue_type
, tissue_data.storage_medium
, tissue_data.misid_status
, tissue_data.collection_date_status
, tissue_data.multi_indivs
, tissue_data.notes
, hormone_sample_data.hsid
, hormone_sample_data.fzdried_date
, hormone_sample_data.sifted_date
, hormone_sample_data.avail_mass_g
, hormone_sample_data.avail_date
, hormone_sample_data.comments;
Table 6.69. Columns in the TISSUES_HORMONES View
| Column | From | Description |
|---|---|---|
| TId | TISSUE_DATA.TId | Identifier for this sample. |
| LocId | TISSUE_DATA.LocId | Identifier for this sample's Institution-Location pair. |
| Institution | LOCATIONS.Institution | Identifier for this sample's locale. |
| Location | LOCATIONS.Location | The current place/position of the sample. |
| LocalId_1 | TISSUE_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #1. |
| LocalId_2 | TISSUE_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #2. |
| UIId | TISSUE_DATA.UIId | Identifier for the source individual, if recorded. |
| PopId | UNIQUE_INDIVS.PopId | Identifier for the population of the source individual, if the source individual is recorded. |
| IndivId | UNIQUE_INDIVS.IndivId | Name/ID of the source individual, if the source individual is recorded. |
| Sname | BIOGRAPH.Sname | Sname of the source individual, if the source individual is recorded and they have an Sname. |
| Name_on_Tube | TISSUE_DATA.Name_on_Tube | Name or ID of the source individual, according to the label on the tube. |
| Collection_Date | TISSUE_DATA.Collection_Date | Date that the sample was collected. |
| Collection_Time | TISSUE_DATA.Collection_Time | Time that the sample was collected. |
| Tissue_Type | TISSUE_DATA.Tissue_Type | The tissue sample type. |
| Storage_Medium | TISSUE_DATA.Storage_Medium | The medium used for storing the sample. |
| Misid_Status | TISSUE_DATA.Misid_Status | The mis-identification status of the sample, if the source individual is recorded. |
| Collection_Date_Status | TISSUE_DATA.Collection_Date_Status | The status of this Collection_Date |
| Multi_Indivs | TISSUE_DATA.Multi_Indivs | Boolean indicating whether this sample has tissue sources from more than one individual. |
| Tissue_Sources | COUNT(TISSUE_SOURCES.*) | The number of rows in TISSUE_SOURCES with this TId. |
| Notes | TISSUE_DATA.Notes | Miscellaneous notes about the sample. |
| HSId | HORMONE_SAMPLE_DATA.HSId | User-generated identifier for the tissue sample. |
| FzDried_Date | HORMONE_SAMPLE_DATA.FzDried_Date | Date the sample was freeze-dried. |
| Sifted_Date | HORMONE_SAMPLE_DATA.Sifted_Date | Date the freeze-dried sample was sifted. |
| Avail_Mass_g | HORMONE_SAMPLE_DATA.Avail_Mass_g | Amount of sample (in g) remaining in the tube, as of the Avail_Date. |
| Avail_Date | HORMONE_SAMPLE_DATA.Avail_Date | Date that the Avail_Mass_g was determined. |
| Comments | HORMONE_SAMPLE_DATA.Comments | Miscellaneous notes/comments about this sample that are relevant only to hormone analysis. |
Because the primary purpose of this view is facilitate working with TISSUE_DATA and HORMONE_SAMPLE_DATA rows simultaneously, this view only allows operations where there is a practical need to operate on the two tables simultaneously[280]. UPDATE is not allowed on TISSUES_HORMONES; there are no needs satisfied nor utility gained from being able to update both tables at once.
Tip
To update data that appear in this view, use the TISSUES or HORMONE_SAMPLES views.
Inserting a row into TISSUES_HORMONES inserts a row into TISSUE_DATA, then a row into HORMONE_SAMPLE_DATA with the same TId. Additional rows may be inserted into TISSUE_LOCAL_IDS, as discussed above.
It is not necessary to provide all of the UIId, PopId, IndivId, and Sname columns, as discussed above.
To indicate a sample's current locale and location, either the LocId column or both the Institution and Location columns must be provided. If all three are provided, the Institution and Location must be equal to the related columns in LOCATIONS for the provided LocId.
The TISSUE_SOURCES column is calculated automatically.
When inserting data, there cannot possibly be any rows in
TISSUE_SOURCES yet, so if a value is
provided for this column it must be
0.
Deleting a row from TISSUES_HORMONES deletes the underlying row from TISSUE_DATA and from HORMONE_SAMPLE_DATA, as expected. Related rows in TISSUE_LOCAL_IDS and TISSUE_SOURCES, if any, are also deleted.
Contains one row for every row in QUAD_DATA.
This view is useful for querying and maintaining the QUAD_DATA table when it is convenient to have X and Y coordinates as separate values instead of geospatial points.
Figure 6.146. Query Defining the QUADS View
SELECT quad_data.quad AS quad
, ST_X(quad_data.xyloc) AS x
, ST_Y(quad_data.xyloc) AS y
, quad_data.aerial AS aerial
FROM quad_data;
Table 6.70. Columns in the QUADS View
| Column | From | Description |
|---|---|---|
| Quad | QUAD_DATA.Quad | Identifier of the map quadrant. |
| X | ST_X(QUAD_DATA.XYLoc) | X coordinate of the XYLoc -- X coordinate of the centroid of the map quadrant. |
| Y | ST_Y(QUAD_DATA.XYLoc) | Y coordinate of the XYLoc -- Y coordinate of the centroid of the map quadrant. |
| Aerial | AERIALS.Aerial | Code indicating the aerial photo in which the map quadrant is located. |
Contains one row for every row in SWERB_DATA.
This view is useful for querying the SWERB_DATA table because it unifies data that is distributed throughout the various SWERB tables. It is also useful when it is convenient to have X and Y or longitude and latitude coordinates as separate values instead of geospatial points.
Note
For more information on the X and Y coordinates see the description of the columns in the underlying tables, see the SWERB Data overview, and see the Amboseli Baboon Research Project Monitoring Guide.
Figure 6.148. Query Defining the SWERB View
SELECT swerb_data.swid AS swid
, swerb_departs_data.did AS did
, swerb_departs_data.date AS date
, swerb_data.time AS time
, swerb_bes.beid AS beid
, swerb_bes.focal_grp AS focal_grp
, swerb_bes.seq AS seq
, swerb_data.event AS event
, swerb_data.seen_grp AS seen_grp
, swerb_data.lone_animal AS lone_animal
, swerb_data.quad AS quad
, CASE
WHEN swerb_data.quad IS NOT NULL
THEN 'quad'
WHEN swerb_data.xyloc IS NULL
THEN 'n/a'
ELSE 'gps'
END AS xysource
, COALESCE(ST_X(swerb_data.xyloc), ST_X(quad_data.xyloc))
AS x
, COALESCE(ST_Y(swerb_data.xyloc), ST_Y(quad_data.xyloc))
AS y
, COALESCE(ST_X(ST_TRANSFORM(swerb_data.xyloc, 4326))
, ST_X(ST_TRANSFORM(quad_data.xyloc, 4326)))
AS long
, COALESCE(ST_Y(ST_TRANSFORM(swerb_data.xyloc, 4326))
, ST_Y(ST_TRANSFORM(quad_data.xyloc, 4326)))
AS lat
, swerb_data.altitude AS altitude
, swerb_data.pdop AS pdop
, swerb_data.accuracy AS accuracy
, swerb_data.subgroup AS subgroup
, swerb_data.ogdistance AS ogdistance
, swerb_data.gps_datetime AS gps_datetime
, swerb_data.garmincode AS garmincode
, swerb_data.predator AS predator
, swerb_loc_data.loc AS loc
, swerb_loc_data.adcode AS adcode
, adcodes.adn AS adn
, swerb_loc_data.loc_status AS loc_status
, swerb_loc_data.adtime AS adtime
, ST_X(swerb_loc_gps.xyloc) AS second_x
, ST_Y(swerb_loc_gps.xyloc) AS second_y
, ST_X(ST_TRANSFORM(swerb_loc_gps.xyloc, 4326)) AS second_long
, ST_Y(ST_TRANSFORM(swerb_loc_gps.xyloc, 4326)) AS second_lat
, swerb_loc_gps.altitude AS second_altitude
, swerb_loc_gps.pdop AS second_pdop
, swerb_loc_gps.accuracy AS second_accuracy
, swerb_loc_gps.gps_datetime AS second_gps_datetime
, swerb_loc_gps.garmincode AS second_garmincode
, swerb_bes.start AS start
, swerb_bes.btimeest AS btimeest
, swerb_bes.bsource AS bsource
, swerb_bes.stop AS stop
, swerb_bes.etimeest AS etimeest
, swerb_bes.esource AS esource
, swerb_bes.is_effort AS is_effort
, swerb_departs_gps.gps AS gps
, swerb_bes.notes AS notes
FROM swerb_data
LEFT OUTER JOIN quad_data
ON (quad_data.quad = swerb_data.quad)
JOIN swerb_bes
ON (swerb_bes.beid = swerb_data.beid)
JOIN swerb_departs_data
ON (swerb_departs_data.did = swerb_bes.did)
LEFT OUTER JOIN swerb_departs_gps
ON (swerb_departs_gps.did = swerb_bes.did)
LEFT OUTER JOIN swerb_loc_data
ON (swerb_loc_data.swid = swerb_data.swid)
LEFT OUTER JOIN adcodes ON (adcodes.adcode = swerb_loc_data.adcode)
LEFT OUTER JOIN swerb_loc_gps
ON (swerb_loc_gps.swid = swerb_loc_data.swid);
Table 6.71. Columns in the SWERB View
| Column | From | Description |
|---|---|---|
| SWId | SWERB_DATA.SWId | Identifier of the record of the SWERB event. |
| DId | SWERB_DEPARTS_DATA.DId | Identifier of the record of departure from camp of the observation team which recorded the SWERB event. |
| Date | SWERB_DEPARTS_DATA.Date | The date of the observation. |
| Time | SWERB_DATA.Time | The time of the observation. |
| BEId | SWERB_BES.BEId | Identifier of the bout of uninterrupted observation of the focal group containing observed SWERB event. |
| Focal_grp | SWERB_BES.Focal_grp | Identifier of the focal group, the group the observation team set out to watch. |
| Seq | SWERB_BES.Seq | A sequence number indicating the ordering of the bouts of uninterrupted observation of each group each day -- ordering of BEId per Focal_grp per Date. |
| Event | SWERB_DATA.Event | Code identifying the type of SWERB event observed. |
| Seen_grp | SWERB_DATA.Seen_grp | Identifier of the observed group. |
| Lone_Animal | SWERB_DATA.Lone_Animal | Sname of the observed lone animal or NULL
when either there is none or an unknown lone male
was observed. |
| Quad | SWERB_DATA.Quad | The code identifying the map quadrant locating the recorded event. |
| XYSource |
| The source of the view's X and Y columns:
The XYSource column values
|
| X | QUAD_DATA.XYLoc or SWERB_DATA.XYLoc | Whatever X geolocation coordinate exists. |
| Y | QUAD_DATA.XYLoc or SWERB_DATA.XYLoc | Whatever Y geolocation coordinate exists. |
| Long | QUAD_DATA.XYLoc or SWERB_DATA.XYLoc | Whatever longitude coordinate exists. |
| Lat | QUAD_DATA.XYLoc or SWERB_DATA.XYLoc | Whatever latitude coordinate exists. |
| Altitude | SWERB_DATA.Altitude | The altitude of the SWERB event. |
| PDOP | SWERB_DATA.PDOP | The PDOP of the SWERB event. |
| Accuracy | SWERB_DATA.Accuracy | Accuracy of the SWERB event. |
| Subgroup | SWERB_DATA.Subgroup | Whether or not the SWERB event pertains to a subgroup. |
| OGDistance | SWERB_DATA.Ogdistance | The distance to the non-focal group (the Seen_grp) at the time the waypoint was taken. |
| GPS_Datetime | SWERB_DATA.GPS_Datetime | The timestamp, the date and time, automatically recorded by the GPS unit when the waypoint was entered into the GPS. |
| Garmincode | SWERB_DATA.Garmincode | The raw data entered by the observer recording the SWERB event. |
| Predator | SWERB_DATA.Predator | The type of predator seen, or NULL when
there is none. |
| Loc | SWERB_LOC_DATA.Loc | Identifier of the related landscape feature, the SWERB_GWS.Loc. |
| ADcode | SWERB_LOC_DATA.ADcode | The code denoting the relationship between the group and the landscape feature. |
| ADN | ADCODES.ADN | Whether the relationship between the group
and the landscape feature is an ascent into a
sleeping grove
(A), a descent
from a sleeping grove
(D), or neither
(N). |
| Loc_Status | SWERB_LOC_DATA.Loc_Status | Code representing the status of the team's observation of the indicated landscape feature. |
| ADtime | SWERB_LOC_DATA.ADtime | Median time of group descent from or ascent into the sleeping grove. |
| Second_X | SWERB_LOC_GPS.XYLoc | The X geolocation coordinate of the 2nd waypoint entry required by the data entry protocol. |
| Second_Y | SWERB_LOC_GPS.XYLoc | The Y geolocation coordinate of the 2nd waypoint entry required by the data entry protocol. |
| Second_Long | SWERB_LOC_GPS.XYLoc | The longitude coordinate of the 2nd waypoint entry required by the data entry protocol. |
| Second_Lat | SWERB_LOC_GPS.XYLoc | The latitude coordinate of the 2nd waypoint entry required by the data entry protocol. |
| Second_Altitude | SWERB_LOC_GPS.Altitude | The altitude of the 2nd waypoint entry required by the data entry protocol. |
| Second_PDOP | SWERB_LOC_GPS.PDOP | The PDOP of the 2nd waypoint entry required by the data entry protocol. |
| Second_Accuracy | SWERB_LOC_GPS.Accuracy | Accuracy of the 2nd waypoint entry required by the data entry protocol. |
| Second_GPS_Datetime | SWERB_LOC_GPS.GPS_Datetime | The timestamp, the date and time, automatically recorded by the GPS unit when the 2nd waypoint required by the data entry protocol was entered into the GPS. |
| Second_Garmincode | SWERB_LOC_GPS.Garmincode | The raw data entered by the observer in the 2nd waypoint entry required by the data entry protocol when recording the SWERB event. |
| Start | SWERB_BES.Start | The time the bout of observation began. |
| Btimeest | SWERB_BES.Btimeest | Whether or not the start time of the bout of observation was estimated. |
| Bsource | SWERB_BES.Bsource | The source of the bout start time value. |
| Stop | SWERB_BES.Stop | The time the bout of observation ended. |
| Etimeest | SWERB_BES.Etimeest | Whether or not the end time of the bout of observation was estimated. |
| Esource | SWERB_BES.Esource | The source of the bout end time value. |
| Is_Effort | SWERB_BES.Is_Effort | Whether or not the bout of observation counts toward total observer effort. |
| GPS | SWERB_DEPARTS_GPS.GPS | Identifier of the GPS device used to record the SWERB event. |
| Notes | SWERB_BES.Notes | Notes on the bout of observation. |
Contains one row for every row in SWERB_DATA.
This view is useful when it is convenient to have X and Y coordinates as separate values instead of geospatial points. For this reason is it also useful when maintaining the SWERB_DATA table. Users querying the data may prefer the SWERB view.
Figure 6.150. Query Defining the SWERB_DATA_XY View
SELECT swerb_data.swid AS swid
, swerb_data.beid AS beid
, swerb_data.seen_grp AS seen_grp
, swerb_data.lone_animal AS lone_animal
, swerb_data.event AS event
, swerb_data.time AS time
, swerb_data.quad AS quad
, ST_X(swerb_data.xyloc) AS x
, ST_Y(swerb_data.xyloc) AS y
, ST_X(ST_TRANSFORM(swerb_data.xyloc, 4326)) AS long
, ST_Y(ST_TRANSFORM(swerb_data.xyloc, 4326)) AS lat
, swerb_data.altitude AS altitude
, swerb_data.pdop AS pdop
, swerb_data.accuracy AS accuracy
, swerb_data.subgroup AS subgroup
, swerb_data.ogdistance AS ogdistance
, swerb_data.gps_datetime AS gps_datetime
, swerb_data.garmincode AS garmincode
, swerb_data.predator AS predator
FROM swerb_data;
Table 6.72. Columns in the SWERB_DATA_XY View
| Column | From | Description |
|---|---|---|
| SWId | SWERB_DATA.SWId | Identifier of the record of the SWERB event. |
| BEId | SWERB_BES.BEId | Identifier of the bout of uninterrupted observation of the focal group containing observed SWERB event. |
| Seen_grp | SWERB_DATA.Seen_grp | Identifier of the observed group. |
| Lone_Animal | SWERB_DATA.Lone_Animal | Sname of the observed lone male or NULL
when either there is none or an unknown lone male
was observed. |
| Event | SWERB_DATA.Event | Code identifying the kind of SWERB event observed. |
| Time | SWERB_DATA.Time | The time of the observation. |
| Quad | SWERB_DATA.Quad | The code identifying the map quadrant locating the recorded event. |
| X | ST_X(SWERB_DATA.XYLoc) | X coordinate of the XYLoc -- X coordinate of the event. |
| Y | ST_Y(SWERB_DATA.XYLoc) | Y coordinate of the XYLoc -- Y coordinate of the event. |
| Long | ST_X(ST_TRANSFORM(SWERB_DATA.XYLoc, 4326)) | Longitude of the XYLoc -- longitude of the event. |
| Lat | ST_Y(ST_TRANSFORM(SWERB_DATA.XYLoc, 4326)) | Latitude of the XYLoc -- latitude of the event. |
| Altitude | SWERB_DATA.Altitude | The altitude of the SWERB event. |
| PDOP | SWERB_DATA.PDOP | The PDOP of the SWERB event. |
| Accuracy | SWERB_DATA.Accuracy | Accuracy of the SWERB event. |
| Subgroup | SWERB_DATA.Subgroup | Whether or not the SWERB event pertains to a subgroup. |
| OGDistance | SWERB_DATA.Ogdistance | The distance to the non-focal group where the SWERB event takes place. |
| GPS_Datetime | SWERB_DATA.GPS_Datetime | The timestamp, the date and time, automatically recorded by the GPS unit when the waypoint was entered into the GPS. |
| Garmincode | SWERB_DATA.Garmincode | The raw data entered by the observer recording the SWERB event. |
| Predator | SWERB_DATA.Predator | The type of predator seen, or NULL when
there is none. |
Inserting a row into SWERB_DATA_XY inserts a row into SWERB_DATA as expected.
Updating the SWERB_DATA_XY view updates the SWERB_DATA table as expected.
Deleting a row from SWERB_DATA_XY deletes a row from SWERB_DATA as expected.
Contains one row for every row in SWERB_DEPARTS_DATA. Each row contains the SWERB_DEPARTS_DATA data and the related SWERB_DEPARTS_GPS row, excepting the geolocation
data which is converted into X and Y coordinates. In those
cases where there is a SWERB_DEPARTS_DATA
row but not a row from SWERB_DEPARTS_GPS the
columns from SWERB_DEPARTS_GPS are
NULL.
This view is useful when downloading departure data for analysis outside of the database, and useful for deleting all information related to specified departures.
Figure 6.152. Query Defining the SWERB_DEPARTS View
SELECT swerb_departs_data.did AS did
, swerb_departs_data.date AS date
, swerb_departs_data.time AS time
, ST_X(swerb_departs_gps.xyloc) AS x
, ST_Y(swerb_departs_gps.xyloc) AS y
, ST_X(ST_TRANSFORM(swerb_departs_gps.xyloc, 4326)) AS long
, ST_Y(ST_TRANSFORM(swerb_departs_gps.xyloc, 4326)) AS lat
, swerb_departs_gps.altitude AS altitude
, swerb_departs_gps.pdop AS pdop
, swerb_departs_gps.accuracy AS accuracy
, swerb_departs_gps.gps AS gps
, swerb_departs_gps.garmincode AS garmincode
FROM swerb_departs_data
LEFT OUTER JOIN swerb_departs_gps
ON (swerb_departs_gps.did = swerb_departs_data.did);
Table 6.73. Columns in the SWERB_DEPARTS View
| Column | From | Description |
|---|---|---|
| Did | SWERB_DEPARTS_DATA.DId | Identifier of the team's departure row. |
| Date | SWERB_DEPARTS_DATA.Date | Date of departure. |
| Time | SWERB_DEPARTS_DATA.Time | Time of the team's departure. |
| X | ST_X(SWERB_DEPARTS_GPS.XYLoc) | X coordinate of the XYLoc -- X coordinate of the point of departure. |
| Y | ST_Y(SWERB_DEPARTS_GPS.XYLoc) | Y coordinate of the XYLoc -- Y coordinate of the point of departure. |
| Long | ST_X(ST_TRANSFORM(SWERB_DEPARTS_GPS.XYLoc, 4326)) | Longitude of the XYLoc -- longitude of the point of departure. |
| Lat | ST_Y(ST_TRANSFORM(SWERB_DEPARTS_GPS.XYLoc, 4326)) | Latitude of the XYLoc -- latitude of the point of departure. |
| Altitude | SWERB_DEPARTS_GPS.Altitude | Altitude at the point of departure. |
| PDOP | SWERB_DEPARTS_GPS.PDOP | Positional Dilution of Precision of the departure's geolocation. |
| Accuracy | SWERB_DEPARTS_GPS.Accuracy | Accuracy of the departure's geolocation expressed as distance in meters. |
| GPS | SWERB_DEPARTS_GPS.GPS | Identifier of the GPS device (GPS_UNITS.GPS) used by the team. |
| Garmincode | SWERB_DEPARTS_GPS.Garmincode | The information manually entered into the waypoint by the observer. |
Inserting a row into SWERB_DEPARTS inserts a row
into SWERB_DEPARTS_DATA and a row into
SWERB_DEPARTS_GPS as expected. Rows are
inserted into SWERB_DEPARTS_GPS when any
of the the relevant columns are present and contain
non-NULL values.
The SWERB_DEPARTS view may be updated and SWERB_DEPARTS_DATA and SWERB_DEPARTS_GPS are (mostly) updated as expected.
Warning
Attempts to update SWERB_DEPARTS_GPS columns when no underlying row exists are silently ignored.
Deleting a row from SWERB_DEPARTS deletes all SWERB data collected by the departing observation team; a row from SWERB_DEPARTS_DATA is deleted along with, if necessary, a row from SWERB_DEPARTS_GPS and multiple related rows from SWERB_OBSERVERS, multiple related rows from SWERB_BES, multiple rows related to these from SWERB_DATA, SWERB_LOC_DATA, and SWERB_LOC_GPS.
Contains one row for every row in SWERB_GW_LOC_DATA.
This view is useful for querying the SWERB_GW_LOC_DATA table because it unifies data that are distributed between the SWERB_GW_LOC_DATA table and the QUAD_DATA table. It is also useful when it is convenient to have X and Y coordinates as separate values instead of geospatial points.
Note
For more information regarding the X and Y coordinates see the description of the columns in the underlying tables, and see the SWERB Data overview.
Figure 6.154. Query Defining the SWERB_GW_LOCS View
SELECT swerb_gw_loc_data.sgwlid AS sgwlid
, swerb_gw_loc_data.loc AS loc
, swerb_gw_loc_data.date AS date
, swerb_gw_loc_data.time AS time
, swerb_gw_loc_data.quad AS quad
, CASE
WHEN swerb_gw_loc_data.xyloc IS NULL
THEN 'quad'
ELSE swerb_gw_loc_data.xysource
END AS xysource
, COALESCE(ST_X(swerb_gw_loc_data.xyloc), ST_X(quad_data.xyloc))
AS x
, COALESCE(ST_Y(swerb_gw_loc_data.xyloc), ST_Y(quad_data.xyloc))
AS y
, COALESCE(ST_X(ST_TRANSFORM(swerb_gw_loc_data.xyloc, 4326))
, ST_X(ST_TRANSFORM(quad_data.xyloc, 4326)))
AS long
, COALESCE(ST_Y(ST_TRANSFORM(swerb_gw_loc_data.xyloc, 4326))
, ST_Y(ST_TRANSFORM(quad_data.xyloc, 4326)))
AS lat
, swerb_gw_loc_data.altitude AS altitude
, swerb_gw_loc_data.pdop AS pdop
, swerb_gw_loc_data.accuracy AS accuracy
, swerb_gw_loc_data.gps AS gps
, swerb_gw_loc_data.notes AS notes
FROM swerb_gw_loc_data
LEFT OUTER JOIN quad_data
ON (quad_data.quad = swerb_gw_loc_data.quad);
Table 6.74. Columns in the SWERB_GW_LOCS View
| Column | From | Description |
|---|---|---|
| SGWLId | SWERB_GW_LOC_DATA.SGWLId | Identifier of the observation of a grove or waterhole's geolocation. |
| Loc | SWERB_GW_LOC_DATA.Loc | Identifier of the object, the grove or waterhole. |
| Date | SWERB_GW_LOC_DATA.Date | The date of the observation. |
| Time | SWERB_GW_LOC_DATA.Time | The time of the observation. |
| Quad | SWERB_GW_LOC_DATA.Quad | The code identifying the map quadrant containing the grove or waterhole. |
| XYSource |
| The source of the view's X, Y, Long, and Lat
columns. When quad
the source of those columns are the coordinates of
the centroid of the related map quadrant. Otherwise
this is the value of the SWERB_GW_LOC_DATA.XYSource
column. |
| X | QUAD_DATA.XYLoc or SWERB_GW_LOC_DATA.XYLoc | Whatever X geolocation coordinate exists. |
| Y | QUAD_DATA.XYLoc or SWERB_GW_LOC_DATA.XYLoc | Whatever Y geolocation coordinate exists. |
| Long | ST_X(ST_TRANSFORM(QUAD_DATA.XYLoc or SWERB_GW_LOC_DATA.XYLoc, 4326)) | Whatever longitude coordinate exists. |
| Lat | ST_Y(ST_TRANSFORM(QUAD_DATA.XYLoc or SWERB_GW_LOC_DATA.XYLoc, 4326)) | Whatever latitude coordinate exists. |
| Altitude | SWERB_GW_LOC_DATA.Altitude | The altitude of the object, the grove or waterhole. |
| PDOP | SWERB_GW_LOC_DATA.PDOP | The PDOP of the object's geolocation. |
| Accuracy | SWERB_GW_LOC_DATA.Accuracy | Accuracy of the object's geolocation. |
| GPS | SWERB_GW_LOC_DATA.GPS | Identifier of the GPS unit (GPS_UNITS) used to take the measurement. |
| Notes | SWERB_GW_LOC_DATA.Notes | Notes on the measurement. |
Contains one row for every row in SWERB_GW_LOC_DATA.
This view is useful when it is convenient to have X and Y coordinates as separate values instead of geospatial points. For this reason is it also useful when maintaining the SWERB_GW_LOC_DATA table. Users querying the view may prefer the SWERB_GW_LOCS view.
Figure 6.156. Query Defining the SWERB_GW_LOC_DATA_XY View
SELECT swerb_gw_loc_data.sgwlid AS sgwlid
, swerb_gw_loc_data.loc AS loc
, swerb_gw_loc_data.date AS date
, swerb_gw_loc_data.time AS time
, swerb_gw_loc_data.quad AS quad
, swerb_gw_loc_data.xysource AS xysource
, ST_X(swerb_gw_loc_data.xyloc) AS x
, ST_Y(swerb_gw_loc_data.xyloc) AS y
, ST_X(ST_TRANSFORM(swerb_gw_loc_data.xyloc, 4326)) AS long
, ST_Y(ST_TRANSFORM(swerb_gw_loc_data.xyloc, 4326)) AS lat
, swerb_gw_loc_data.altitude AS altitude
, swerb_gw_loc_data.pdop AS pdop
, swerb_gw_loc_data.accuracy AS accuracy
, swerb_gw_loc_data.gps AS gps
, swerb_gw_loc_data.notes AS notes
FROM swerb_gw_loc_data;
Table 6.75. Columns in the SWERB_GW_LOC_DATA_XY View
| Column | From | Description |
|---|---|---|
| SGWLId | SWERB_GW_LOC_DATA.SGWLId | Identifier of the observation that geolocated the object, the grove or waterhole. |
| Loc | SWERB_GW_LOC_DATA.Loc | Identifier of the object, the grove or waterhole, that is located. |
| Date | SWERB_GW_LOC_DATA.Date | The date of the observation. |
| Time | SWERB_GW_LOC_DATA.Time | The time of the observation. |
| Quad | SWERB_GW_LOC_DATA.Quad | The code identifying the map quadrant containing the observed object, the grove or waterhole. |
| X | ST_X(SWERB_GW_LOC_DATA.XYLoc) | X coordinate of the XYLoc -- X coordinate of the object. |
| Y | ST_Y(SWERB_GW_LOC_DATA.XYLoc) | Y coordinate of the XYLoc -- Y coordinate of the object. |
| Long | ST_X(ST_TRANSFORM(SWERB_GW_LOC_DATA.XYLoc, 4326)) | Longitude of the XYLoc -- longitude of the object. |
| Lat | ST_Y(ST_TRANSFORM(SWERB_GW_LOC_DATA.XYLoc, 4326)) | Latitude of the XYLoc -- latitude of the object. |
| Altitude | SWERB_GW_LOC_DATA.Altitude | The altitude of the object. |
| PDOP | SWERB_GW_LOC_DATA.PDOP | The PDOP of the geolocation. |
| Accuracy | SWERB_GW_LOC_DATA.Accuracy | Accuracy of the SWERB geolocation. |
| GPS | SWERB_GW_LOC_DATA.GPS | The code identifying the GPS unit (GPS_UNITS) used to take the observation. |
| Notes | SWERB_GW_LOC_DATA. Notes | Notes on the observation. |
Inserting a row into SWERB_GW_LOC_DATA_XY inserts a row into SWERB_GW_LOC_DATA as expected.
Updating the SWERB_GW_LOC_DATA_XY view updates the SWERB_GW_LOC_DATA table as expected.
Deleting a row from SWERB_GW_LOC_DATA_XY deletes a row from SWERB_GW_LOC_DATA as expected.
Contains one row for every row in SWERB_LOC_GPS.
This view is useful when it is convenient to have X and Y coordinates as separate values instead of geospatial points. For this reason is it also useful when querying and maintaining the SWERB_LOC_GPS table.
Figure 6.158. Query Defining the SWERB_LOC_GPS_XY View
SELECT swerb_loc_gps.swid AS swid
, ST_X(swerb_loc_gps.xyloc) AS x
, ST_Y(swerb_loc_gps.xyloc) AS y
, ST_X(ST_TRANSFORM(swerb_loc_gps.xyloc, 4326)) AS long
, ST_Y(ST_TRANSFORM(swerb_loc_gps.xyloc, 4326)) AS lat
, swerb_loc_gps.altitude AS altitude
, swerb_loc_gps.pdop AS pdop
, swerb_loc_gps.accuracy AS accuracy
, swerb_loc_gps.gps_datetime AS gps_datetime
, swerb_loc_gps.garmincode AS garmincode
FROM swerb_loc_gps;
Table 6.76. Columns in the SWERB_LOC_GPS_XY View
| Column | From | Description |
|---|---|---|
| SWId | SWERB_LOC_GPS.SWId | Identifier of the GPS information involving an observation of a group at a particular time at a particular grove or waterhole. Also the SWERB_DATA.SWId value and the SWERB_LOC_DATA.SWId value |
| X | ST_X(SWERB_LOC_GPS.XYLoc) | X coordinate of the XYLoc -- X coordinate of the group. |
| Y | ST_Y(SWERB_LOC_GPS.XYLoc) | Y coordinate of the XYLoc -- Y coordinate of the group. |
| Long | ST_X(ST_TRANSFORM(SWERB_LOC_GPS.XYLoc, 4326)) | Longitude of the XYLoc -- Longitude of the group. |
| Lat | ST_Y(ST_TRANSFORM(SWERB_LOC_GPS.XYLoc, 4326)) | Latitude of the XYLoc -- latitude of the group. |
| Altitude | SWERB_LOC_GPS.Altitude | The altitude of the group. |
| PDOP | SWERB_LOC_GPS.PDOP | The PDOP of the geolocation. |
| Accuracy | SWERB_LOC_GPS.Accuracy | Accuracy of the SWERB geolocation. |
| GPS_Datetime | SWERB_LOC_GPS.GPS_Datetime | The date and time recorded by the GPS unit. |
| Garmincode | SWERB_LOC_GPS. Garmincode | The information manually entered into the waypoint by the observer. |
Inserting a row into SWERB_LOC_GPS_XY inserts a row into SWERB_LOC_GPS as expected.
Updating the SWERB_LOC_GPS_XY view updates the SWERB_LOC_GPS table as expected.
Deleting a row from SWERB_LOC_GPS_XY deletes a row from SWERB_LOC_GPS as expected.
Contains one row for every row in SWERB_LOC_DATA.
This view is useful for querying the SWERB_LOC_DATA table because makes explicit whether or not the landscape feature involves descent from or ascent into a sleeping grove.
Figure 6.160. Query Defining the SWERB_LOCS View
SELECT swerb_loc_data.swid AS swid
, swerb_loc_data.loc AS loc
, swerb_loc_data.adcode AS adcode
, adcodes.adn AS adn
, swerb_loc_data.loc_status AS loc_status
, swerb_loc_data.adtime AS time
FROM swerb_loc_data
JOIN adcodes ON (adcodes.adcode = swerb_loc_data.adcode);
Table 6.77. Columns in the SWERB_LOCS View
| Column | From | Description |
|---|---|---|
| SWId | SWERB_LOC_DATA.SWId | Identifier of the placement of the group at the landscape feature and of the related of the SWERB event, the SWERB_DATA.SWId. |
| Loc | SWERB_LOC_DATA.Loc | Identifier of the related landscape feature, the SWERB_GWS.Loc. |
| ADcode | SWERB_LOC_DATA.ADcode | The code denoting the relationship between the group and the landscape feature. |
| ADN | ADCODES.ADN | Whether the relationship between the group
and the landscape feature is an ascent into a
sleeping grove
(A), a descent
from a sleeping grove
(D), or neither
(N). |
| Loc_Status | SWERB_LOC_DATA.Loc_Status | Code representing the status of the team's observation of the indicated landscape feature. |
| ADtime | SWERB_LOC_DATA.ADtime | Median time of group descent from or ascent into the sleeping grove. |
This view returns no rows, it is used only to upload
data into the SWERB portion of Babase. Attempting to
SELECT rows from this view will raise an
error.
This view exists instead of a custom upload program.
The SWERB_UPLOAD view uses
G as the value for SWERB_BES.Bsource and
SWERB_BES.Esource in the SWERB_BES rows it inserts, unless a different
value is provided in this view's Source column.
Whenever the SWERB_UPLOAD view obtains a SWERB_DATA.Time from the GPS unit (the SWERB_DATA.GPS_Datetime value) instead of from operator entry (the SWERB_DATA.Garmincode value) the seconds portion of the timestamp is discarded.
When a median ascent/descent time is entered into the
GPS unit by the observer the SWERB_UPLOAD program uses the
values A and
D for the ascent and
descent SWERB_LOC_DATA.ADcode value,
respectively.[281] When a drinking event is recorded the SWERB_UPLOAD
view uses the value N
for the SWERB_LOC_DATA.ADcode. (See ADCODES: Special Values.)
When SWERB_UPLOAD
encounters a line which records a drinking event and the
immediately preceding line (or pair of lines in the case of
beginning of observation) is an observation of a subgroup and
the line immediately following (or pair of lines in the case
of end of observation) is an observation of a subgroup then
the SWERB_DATA.Subgroup of the drinking event will be
set to TRUE -- the drinking event will be recorded as that
of a subgroup.[282] When considering whether the preceding and
subsequent lines are of subgroups lines representing drinking
events and lines representing observations of lone animals and
other groups are ignored.
When SWERB_UPLOAD encounters a non-focal group
observation that is not part of any bout of observation it
automatically creates a bout of observation to contain the
non-focal group observation. The created bout of observation
has as its focal the unknown group
(9.0). It begins and ends at the
time of the non-focal group observation and so has a duration
of 0 minutes. It is also marked as a bout of observation
which should not count toward observer effort (SWERB_BES.Is_Effort is
FALSE). Aside from the begin and end rows, the only SWERB
observation (the only SWERB_DATA) row
belonging to the bout is the non-focal group
observation.
The format of the data that is inserted into the SWERB_UPLOAD view is complex. This section provides an overview and remarks on unusual features[283] and the tasks required of the data entry manager to convert the raw SWERB data into an uploadable format. The description of the SWERB_UPLOAD view in the following sections describes how the various uploaded columns map into the columns of Babase's tables. Because, excepting variances described in this section, the uploaded data comes directly from the GPS units used to collect SWERB data the reader should rely on the description of the SWERB data collection protocol in the Amboseli Baboon Research Project Monitoring Guide for a complete description of the data format.[284]
Each upload into the SWERB_UPLOAD view must consist of the data collected on a single GPS unit by a single observation team during the course of a single day.
The data is uploaded as a collection of lines containing tab-delimited text. Each line represents a waypoint recorded by the operator. The lines are expected to be in chronological order, the first line being the first waypoint recorded and the last line being the last, with the exceptions that the lines contriving any one bout of observation of any one (sub)group must be contiguous and that the begin and end lines which record the sleeping grove must immediately precede the begin and end lines which record the descent or ascent time.[285] Consequently the following constraints are imposed on the data: the first line(s) must record the observation team's departure from camp; the lines representing a bout of observation must be contiguous; the line denoting the beginning of a bout of observation must precede all of the bout's other lines; the line denoting the end of a bout of observation must follow all of the bout's other lines; the line recording the median descent time, when present, must immediately follow the line denoting beginning of the bout of observation and the previous night's sleeping grove; the line recording the median ascent time, when present, must immediately follow the line denoting end of the bout of observation and the night's sleeping grove; in those cases where a group utilizes more than one sleeping grove the sleeping grove information must consist of contiguous pairs of lines (as just described) with no intervening lines of another sort; notwithstanding anything to the contrary above, lines denoting observations of the non-focal group may appear at any point after the lines representing departure from camp.[286]
Note
When there is more than one line representing departure of the team from camp the only GPS information recorded in SWERB_DEPARTS_GPS is that of the first departure line. The GPS information (XY coordinates, altitude, pdop, timestamp, etc.) recorded in successive departure rows is discarded; successive departure lines serve only to supply additional observers and their roles for insertion into the SWERB_OBSERVERS table.
The first 2 lines of the uploaded file are required to
be departure lines, lines which record information about the
departing observation team. The first line must begin with
a D, it lists the
observers. The initials supplied on this first line control
the SWERB_OBSERVERS.Role value used, the value used
being the referenced OBSERVERS.SWERB_Observer_Role column's value.
The second line must begin with
DD,
it lists the drivers. The initials supplied on the second
line control the SWERB_OBSERVERS.Role value used, the value used
being the referenced OBSERVERS.SWERB_Driver_Role column's
value.
It is an error if all the lines representing departure from camp indicate the use of more than one GPS unit. Each data upload into SWERB_DEPARTS must come from a single GPS unit.
It is an error if observer codes cannot be unambiguously interpreted in the departure lines. This means that when there is an observer code which is shorter and match in its entirety the beginning of other observer codes then none of these observer codes, neither the shorter nor the longer, can be reliably used in SWERB departure waypoints.[287] If there is ambiguity the offending observer code must be manually removed from the departure line and manually inserted into SWERB_OBSERVERS after uploading the data file.
When the field team records coordinates for the start or stop of a bout of observation but somehow fails to record the time, then that time must be estimated by the data entry staff and included in the Description. In this case the columns Timeest and Source must be added to the data file by the data manager. The data manager should supply values for these columns only in the begin and end lines.
Note
The SWERB_LOC_DATA.ADcode value is sometimes
obtained directly from the GPS waypoint data (the Name
column) entered by the observer, from the second begin/end
line recording median ascent/descent time. This occurs
when, for whatever reason, the operator does not record a
time following after entering the letters
MAT or
MAT. Whatever is entered in
place of a time (which is required to be entered as
4 digits), is used for the SWERB_LOC_DATA.ADcode value.
When the field team fails to record the start or stop
of a bout of observation at all, the data manager needs to
create one. In these cases, coordinates cannot be
estimated, but the start/stop time may be known or estimated
from other data. However, when a date and time are provided
(when Description is not
NULL), it is normally a rule that coordinates must also be
provided (Position cannot be
NULL). To manage this conflict, the boolean column BE_Has_Coords is used. When
FALSE, the Position must be
NULL and the Description is
allowed to be non-NULL[288]. When BE_Has_Coords is TRUE or NULL
and the Description is not
NULL, the Position cannot
be NULL, as usual.
The BE_Has_Coords
column is only used for begins and ends of observation
bouts. This column must be NULL for all other rows.
The data collection protocols require that each observation team record ascent and descent times and groves for their first and last observations of each group for the day. When more than one observation team observes a single group on a given day then the data manager must choose which observation team's ascent/descent information is to be used.[289]The unused ascent and descent information must be removed from the uploaded data. This requires removing the grove from the end of the waypoint text, in the Name column, and deleting the line denoting median descent/ascent time. This should leave a single beginning of observation/end of observation line in the place within the file from which the sleeping grove ascent/descent information has been purged.
The SWERB_UPLOAD view treats a leading
P character before
grove codes written into the uploaded begin and end lines as
an indication that the sleeping grove is probable (SWERB_LOC_DATA.Loc_Status is
P) unless the result of
removing the leading
P produces a code
which is not in SWERB_GWS as a grove. In
this case the leading
P is considered
part of the grove code.
Although the field operators enter information after
the B and E codes in
those waypoints recording the beginning and ending of bouts
of observation that do not denote
descent from or ascent into sleeping groves, the
SWERB_UPLOAD view is unable to process this additional
information. The data manager must remove this information
from those begin and end lines that occur when observation
of the focal group is interrupted for some reason.
When the field team records secondary ascents or
descents, when there are subgroups and more than one begin
or end is recorded for the group, the data managers must add
additional lines to the uploaded data to convert these
“extra” begins or ends into independent bouts
of observation[290]. All of these lines, the original secondary
ascent or descent lines and the additional lines added by
the data manager, must be marked with a TRUE value in a
Secondary_AD column. (This
column will also have to be added by the data
manager.)
The bout of observation created for the secondary ascent or descent must consist only of begin and end rows, no other kinds of observation are allowed. The SWERB_UPLOAD view will generate an error if other kinds of observations are interspersed between the begin and end rows of a secondary ascent or descent.[291]
Secondary ascents and descents must occur during a
regular bout of observation -- uploaded rows with a TRUE
Secondary_AD value must be
preceded by non-secondary begin rows and followed by
non-secondary end rows.
Caution
Although row-wise ordering of secondary bouts of observation is enforced by SWERB_UPLOAD there is no enforcement of time-wise ordering.
Lone animal sightings must be flagged as such in the SWERB_UPLOAD.Lone_Animal column. The sex of the individual must match the sex indicated in the SWERB_UPLOAD.Lone_Animal column.
The SWERB_UPLOAD view looks up the the sname for lone
animal entered as part of the garmincode in the Unksname column of the UNKSNAMES table. If found and the related UNKSNAMES.Lonemale
value is M the sname of the lone
male is stored in SWERB_DATA.Lone_Animal as a NULL.
Note
As usual not all columns need be present in the uploaded data file and, while the column headings are significant, the order of the columns is not. In particular it is expected that older data using the quad coordinate system will use the Quad column in place of the Position column.
Note
Many columns are may be included in the uploaded data but are ignored. This is to reduce the amount of data manipulation which the data manager must perform on the raw data downloaded from the GPS units.
It is an error to include values in both the Description and the Date columns on the same line.
The geographic coordinates for each row are recorded
in the Position column. As
discussed elsewhere, the provided
coordinates may use WGS
1984 UTM Zone 37South coordinates or longitude and
latitude via the WGS
1984 2D CRS, but the location will be
stored in its respective table as a WGS
1984 UTM Zone 37South location. When
a coordinate in longitude or latitude converts to a UTM
coordinate with too many digits after the decimal, the
converted value is rounded to the nearest
0.1.
Because the coordinates in the Position column may be from either system, this view uses the provided Position to guess which system is being used. When the provided value begins with 37 M, the coordinates are assumed to be from the WGS 1984 UTM Zone 37South system. The expected format of those data is “37 M” space [X-coordinate] space [Y-coordinate]. The XY units are in meters and are always positive. When the provided value begins with 37., the coordinates are presumed to be from the WGS 1984 2D CRS. The expected format of those data is [longitude] space [latitude]. The units are decimal degrees, with a positive longitude and a negative latitude.
Figure 6.162. Query Defining the SWERB_UPLOAD View
SELECT NULL::TEXT AS header
, NULL::TEXT AS name
, NULL::TEXT AS description
, NULL::TEXT AS type
, NULL::TEXT AS position
, NULL::TEXT AS altitude
, NULL::TEXT AS depth
, NULL::TEXT AS proximity
, NULL::TEXT AS display_mode
, NULL::TEXT AS color
, NULL::TEXT AS symbol
, NULL::TEXT AS facility
, NULL::TEXT AS city
, NULL::TEXT AS state
, NULL::TEXT AS country
, NULL::TEXT AS pdop
, NULL::TEXT AS accuracy
, NULL::TEXT AS quad
, NULL::TEXT AS date
, NULL::TEXT AS timeest
, NULL::TEXT AS source
, NULL::TEXT AS lone_animal
, NULL::TEXT AS is_effort
, NULL::BOOLEAN AS secondary_ad
, NULL::BOOLEAN AS be_has_coords
, NULL::TEXT AS notes
WHERE _raise_babase_exception(
'Cannot select SWERB_UPLOAD'
|| ': The only use of the SWERB_UPLOAD view is to insert'
|| ' new data into the SWERB portion of babase');
Figure 6.163. Entity Relationship Diagram of the SWERB_UPLOAD View
Table 6.78. Columns in the SWERB_UPLOAD View
| Column | Uploads into | Description |
|---|---|---|
| Header | Data in this column is not inserted into Babase. | A record of which button was pushed on the
GPS unit. It is an error if this value is not
either NULL, as would be the case when the column is
omitted from the uploaded data, or
Waypoint. |
| Name | One or more columns of one or more of SWERB_DEPARTS_DATA, SWERB_DEPARTS_GPS, SWERB_BES, SWERB_DATA, and SWERB_LOC_DATA | The data entered by the field operator when recording the waypoint. The entered text not only supplies data but also drives which tables and columns receive the line's data. See above and the Amboseli Baboon Research Project Monitoring Guide. |
| Description | SWERB_DATA.GPS_Datetime and sometimes also SWERB_DATA.Time, or SWERB_DEPARTS_DATA.Date and SWERB_DEPARTS_DATA. Time, or ignored | This is the timestamp, date and time,
the GPS unit automatically supplies when the
waypoint is taken. With a few exceptions —
camp departure rows, median ascent or descent time
rows, and observations begins or ends whose BE_Has_Coords is If this column is blank ('') or The format of this data is “yyyy/mm/dd space HH:MM”. |
| Type | Ignored | The GPS unit supplies more information about the pressed button in this column. This information is ignored. |
| Position | SWERB_DEPARTS_GPS.XYLoc or SWERB_DATA.XYLoc or ignored | The geolocation coordinates supplied by the GPS unit. In the case of the first of those lines representing departure from camp this is the place of departure. In most of the remainder of this lines this is the location where the data waypoint was taken. See above for when this information is discarded. Also see above for how the system determines which coordinate system is being used. |
| Altitude | SWERB_DEPARTS_GPS.Altitude or SWERB_DATA.Altitude or ignored | The altitude supplied by the GPS unit. In the case of the first of those lines representing departure from camp this is the altitude of the place of departure. In most of the remainder of this lines this is the altitude where the data waypoint was taken. See above for when this information is discarded. The format of this data is numeric, possibly followed by a space and then either the characters “ft” or the character “m”. This value is in meters, unless the characters “ft” are present in which case the value is in feet. The SWERB_UPLOAD view converts feet to meters for storage in the database by multiplying by 0.3048. |
| Depth | Ignored | This information is ignored. |
| Proximity | Ignored | This information is ignored. |
| Display_Mode | Ignored | This information is ignored. |
| Color | Ignored | This information is ignored. |
| Symbol | Ignored | This information is ignored. |
| Facility | Ignored | This information is ignored. |
| City | Ignored | This information is ignored. |
| State | Ignored | This information is ignored. |
| Country | Ignored | This information is ignored. |
| Pdop | SWERB_DEPARTS_GPS.PDOP or SWERB_DATA.PDOP or ignored | The PDOP supplied by the GPS unit. In the case of the first of those lines representing departure from camp this is the PDOP of the departure reading. In most of the remainder of this lines this is the PDOP of the geolocation reading where the data waypoint was taken. See above for when this information is discarded. The format of this data is numeric. |
| Accuracy | SWERB_DEPARTS_GPS.Accuracy or SWERB_DATA.Accuracy or ignored | The accuracy supplied by the GPS unit. In the case of the first of those lines representing departure from camp this is the accuracy of the departure reading. In most of the remainder of this lines this is the accuracy of the geolocation reading where the data waypoint was taken. See above for when this information is discarded. The format of this data is numeric. The units are meters. |
| Quad | SWERB_DATA.Quad or ignored | The quad coordinates of the SWERB waypoint reading. It is an error to supply a quad value for departure lines or for the 2nd begin or end lines. In most of the remainder of this lines this is the location where the data waypoint was taken. See above for when this information is discarded. This data must be a valid QUADS.Quad value. |
| Date | Ignored or SWERB_DEPARTS_DATA.Date | The date of manually recorded SWERB data, data collected before GPS units were put in service. In the case of departure lines this is the departure date. Data supplied manually by the data manager in order that uploaded data conform to the required rules, as when the field team accidentally omits a begin or end record, may use a value on the Date column in lieu of data values in all the columns automatically supplied by the GPS units. In all other lines the value of this column is ignored. The date may be in any format accepted by PostgreSQL. |
| Timeest | Ignored or SWERB_BES.Btimeest or SWERB_BES.Etimeest | Whether the begin or end line was entered by
the data manager and contains an estimated time.
The column must contain no value for those lines
that represent something other than the beginning or
end of a bout of observation. Since the begin/end
time is from the first of the 2 (if 2 are present)
begin/end lines only the first begin and end line
can have an estimated time. The format is any
boolean representation recognized by PostgreSQL.
The empty string, an omitted value, is taken to be
FALSE.
|
| Source | Ignored or SWERB_BES.Bsource or SWERB_BES.Esource | How the data manager obtained the begin or
end time. This must be a SWERB_TIME_SOURCES.Source value, or
blank. This column must contain no value (the empty
string or NULL) for those lines that represent
something other than the beginning or end of a bout
of observation. Since the the begin/end time is
obtained from the first of the 2 (if 2 are present)
begin/end lines only the first begin and end line
can have a source value. The default value, when
the empty string or NULL, is
G. |
| Lone_Animal | Nowhere; controls interpretation of the row | A legal BIOGRAPH.Sex value. When non-NULL and
not empty the row represents a lone animal sighting
and is interpreted as such. When NULL or empty
the row is not a lone animal sighting. |
| Is_Effort | SWERB_BES.Is_Effort | Must be NULL or the empty string unless the
line represents the start of a bout of observation,
and is the first start line when there are more than
one. Whether the bout of observation is to be
counted toward total observer effort. Defaults to
FALSE when not supplied with the first line
representing the beginning of a bout of
observation.[a] |
| Secondary_AD | Nowhere; controls interpretation of the row | A boolean value. All PostgreSQL boolean
representations are accepted. When TRUE the row
represents a secondary ascent or descent, presumably
of a subgroup, and a separate bout of observation
will be created. When FALSE or NULL the row is
not part of a secondary ascent or descent
observation. |
| BE_Has_Coords | Nowhere; controls interpretation of the row | A boolean value. All PostgreSQL boolean
representations are accepted. When FALSE the row
represents a start or stop of observation that was
not recorded in a GPS unit but whose time was
nontheless estimated from other available data.
This is the only case where a Description can be provided
while the Position is
NULL. When TRUE or NULL, the row is not one
of these unusual starts/stops. |
| Notes | SWERB_BES.Notes | Must be NULL or the empty string unless the
line is the first line representing the start of a
bout of observation. Any notes regarding the bout
of observation. Replaces any existing value.
Defaults to NULL and is changed to NULL when the
empty string. |
[a] Lines representing sightings of "other"
(non-focal) groups and lone individuals are not
interpreted as lines beginning observation bouts,
even though these events are recorded in SWERB_BES as independent bouts of
observation. Because of this, an Is_Effort value
cannot be supplied for these events, and therefore
will default to | ||
This view does not currently exist. But it might someday!
This view unifies the temperature minima and maxima from both the manually and electronically collected weather data into a single place. For the manually collected data, this view contains one row for every WREADINGS row with a curated minimum and/or maximum temperature in TEMPMINS and TEMPMAXS. The electronically collected data in DIGITAL_WEATHER are first aggregated into 24-hour periods beginning and ending at 06:00 AM each day, in which the lowest curated" AirTemp_Min and highest curated AirTemp_Max are retained. This view contains one row for each of these 24-hour periods.
Ideally, none of the rows in this view should overlap in time but this is not explicitly forbidden. The system will return a warning when this occurs.
Tip
Data managers can prevent overlaps in time from happening by using the various curated columns to prune overlapping rows.
Figure 6.164. Query Defining the CURATED_TEMPS View
WITH manual AS (SELECT wreadings.wstation
, (SELECT last.wrdaytime
FROM wreadings AS last
WHERE last.wstation = wreadings.wstation
AND last.wrdaytime < wreadings.wrdaytime
AND (EXISTS(SELECT 1 FROM tempmins tm WHERE tm.wrid = wreadings.wrid)
OR
EXISTS(SELECT 1 FROM tempmaxs tm WHERE tm.wrid = wreadings.wrid))
ORDER BY last.wrdaytime DESC
LIMIT 1) AS start
, wreadings.wrdaytime AS stop
, tempmins.tempmin
, tempmaxs.tempmax
FROM wreadings
LEFT JOIN tempmins
ON tempmins.wrid = wreadings.wrid
AND tempmins.curated
LEFT JOIN tempmaxs
ON tempmaxs.wrid = wreadings.wrid
AND tempmaxs.curated
WHERE COALESCE(tempmins.tempmin, tempmaxs.tempmax) IS NOT NULL)
, hourly AS (SELECT digital_weather.wstation
, date_trunc('DAY', timestamp - '7 hours'::interval) AS effective_date
, digital_weather.timestamp
, CASE
WHEN digital_weather.airtemp_min_curated
THEN digital_weather.airtemp_min
ELSE NULL
END AS airtemp_min
, CASE
WHEN digital_weather.airtemp_max_curated
THEN digital_weather.airtemp_max
ELSE NULL
END AS airtemp_max
FROM digital_weather
WHERE digital_weather.airtemp_min_curated
OR digital_weather.airtemp_max_curated
)
, digital AS (SELECT hourly.wstation
, (MIN(hourly.timestamp) - '1 hour'::interval) AS start
, MAX(hourly.timestamp) AS stop
, MIN(hourly.airtemp_min) AS tempmin
, MAX(hourly.airtemp_max) AS tempmax
FROM hourly
GROUP BY hourly.wstation, hourly.effective_date
)
, temps AS (SELECT * FROM manual
UNION
SELECT * FROM digital)
SELECT wstation AS wstation
, start AS start
, stop AS stop
, tempmin AS tempmin
, tempmax AS tempmax
, to_hhmmss(stop - start) AS timespan
, to_hhmmss(start - (LAG(stop) OVER (ORDER BY stop))) AS time_missed
FROM temps;
Figure 6.165. Entity Relationship Diagram of the portion of the CURATED_TEMPS View involving electronically-collected weather data
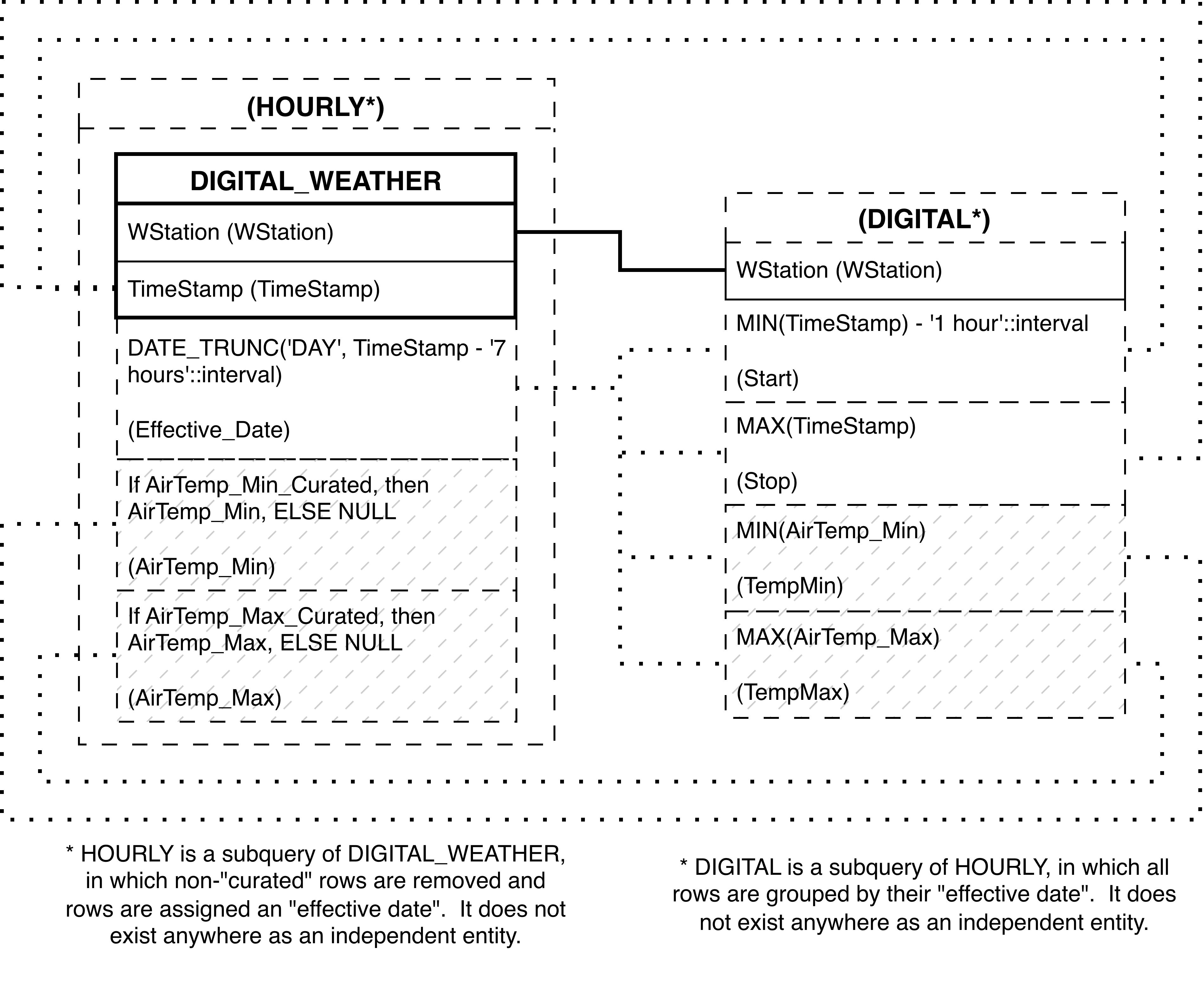 |
Figure 6.166. Entity Relationship Diagram of the portion of the CURATED_TEMPS View involving manually-collected weather data
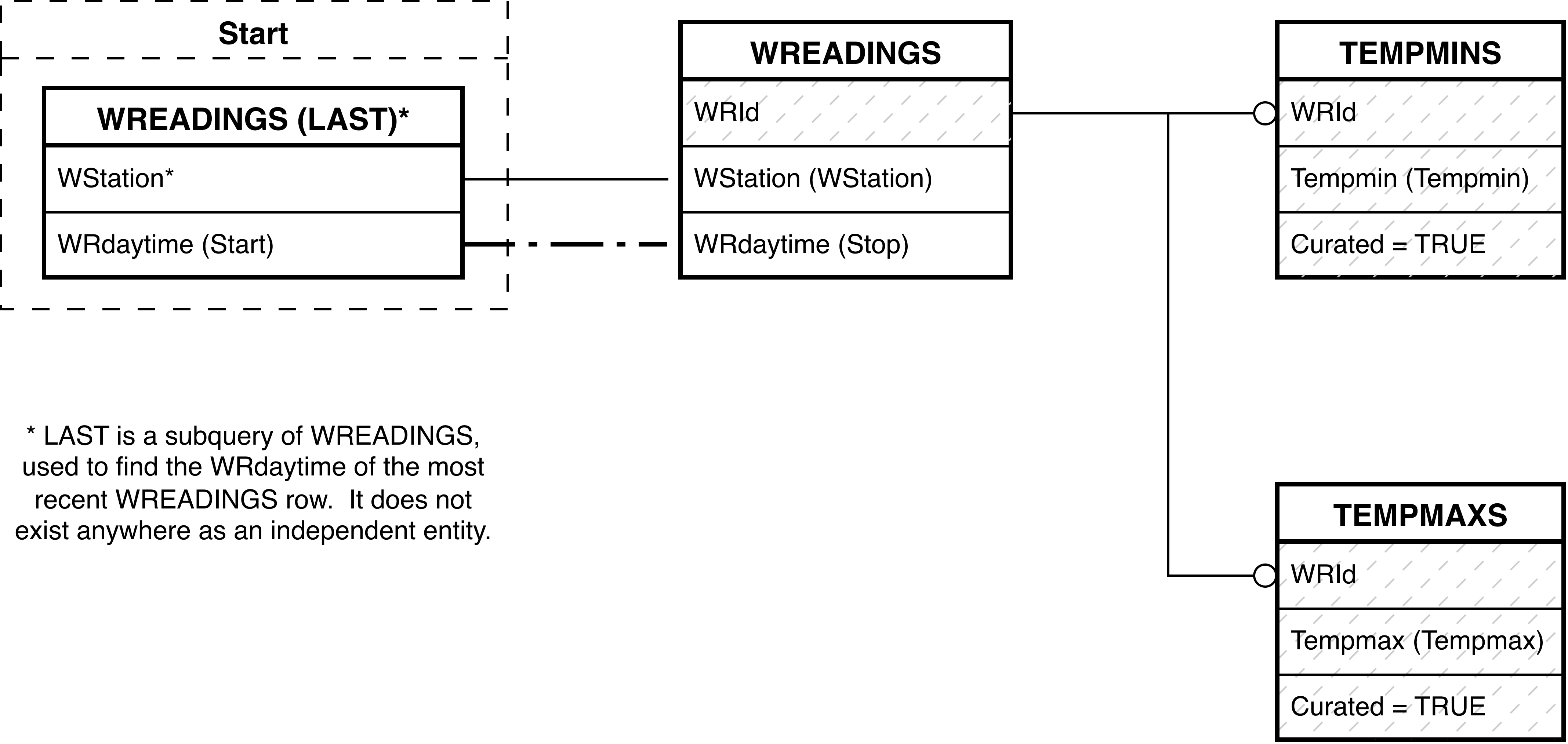 |
Table 6.79. Columns in the CURATED_TEMPS View
| Column | From | Description |
|---|---|---|
| WStation | WREADINGS.Wstation or DIGITAL_WEATHER.WStation | The weather station from which this row's data were collected. |
| Start | WREADINGS.WRdaytime or DIGITAL_WEATHER.TimeStamp | The WRdaytime of
the previous manually collected weather reading
having any curated
temperatures (or NULL, if there is no such reading),
or the earliest (TimeStamp - 1
hour) of all the DIGITAL_WEATHER rows that were
aggregated into the row. |
| Stop | WREADINGS.WRdaytime or DIGITAL_WEATHER.TimeStamp | The WRdaytime of the manually-collected weather reading, or the latest TimeStamp of all the DIGITAL_WEATHER rows that were aggregated into the row. |
| Tempmin | TEMPMINS.Tempmin or DIGITAL_WEATHER.AirTemp_Min | The curated Tempmin (if any) for the manual reading, or the lowest curated AirTemp_Min (if any) of all the DIGITAL_WEATHER rows that were aggregated into the row. |
| Tempmax | TEMPMAXS.Tempmax or DIGITAL_WEATHER.AirTemp_Max | The curated Tempmax (if any) for the manual reading, or the highest curated AirTemp_Max (if any) of all the DIGITAL_WEATHER rows that were aggregated into the row. |
| Timespan | WREADINGS.WRdaytime or DIGITAL_WEATHER.TimeStamp | The time elapsed between this row's Start and Stop, as HH:MM:SS. |
| Time_Missed | WREADINGS.WRdaytime or DIGITAL_WEATHER.TimeStamp | The time elapsed between the chronologically previous row's Stop and this row's Start, as HH:MM:SS. |
Contains one row for every row in WREADINGS. Each row contains the WREADINGS data and the related TEMPMINS, TEMPMAXS, and RAINGAUGES rows. In those cases where there is a
WREADINGS row but not a row from a related
table the columns from the related table are NULL.
This view is useful for the analysis of the manually collected weather data.[292]
Warning
The UNadjusted maximum temperature is not shown in this view. That is, when the original maximum temperature was determined to be spurious and has been adjusted in some way, this view does not provide a way to identify which Tempmax values are and are not adjusted. When this information is important to retain, users should use the TEMPMAXS table.
For more information, see TEMPMAXS and its Historical Note.
Caution
The WRdaytime column indicates when the reading was
recorded, not when the recorded weather
events (e.g. Tempmax, Rain) occurred. The weather data shown
in each row are the weather conditions since the previous
reading — usually covering the day before the WRdaytime.
For example, a Tempmax value in a row with WRdaytime =
2015-07-04 06:00 will usually reflect the
high temperature for 2015-07-03; however, if the previous
WRdaytime was more than 24 hours before the current one, you
will not be able to determine which particular date
experienced that high temperature. The only thing you know is
that that temperature occurred sometime between the current
WRdaytime and the previous one.
It is important to think of each row as the weather conditions since the last reading, not yesterday's weather. Occasionally, one or more days are skipped and thus a single row may represent weather conditions spanning multiple days. Always interpret the values as covering the interval between this reading and the last, not the calendar day shown in WRdaytime.
Tip
If you need a table that converts the WRdaytimes into estimates of rainfall on a daily basis, ask a data manager for help.
Figure 6.167. Query Defining the MIN_MAXS View
SELECT wreadings.wrid AS wrid
, wreadings.wstation AS wstation
, wreadings.wrdaytime AS wrdaytime
, wreadings.estdaytime AS estdaytime
, wreadings.wrperson AS wrperson
, wreadings.wrnotes AS wrnotes
, tempmins.tempmin AS tempmin
, tempmins.curated AS tempmin_curated
, tempmaxs.tempmax AS tempmax
, tempmaxs.curated AS tempmax_curated
, raingauges.rgspan AS rgspan
, raingauges.estrgspan AS estrgspan
, raingauges.rain AS rain
FROM wreadings
LEFT OUTER JOIN tempmins
ON wreadings.wrid = tempmins.wrid
LEFT OUTER JOIN tempmaxs
ON wreadings.wrid = tempmaxs.wrid
LEFT OUTER JOIN raingauges
ON wreadings.wrid = raingauges.wrid;
Table 6.80. Columns in the MIN_MAXS View
| Column | From | Description |
|---|---|---|
| WRid | WREADINGS.WRid | Identifier of the manual weather reading. |
| Wstation | WREADINGS.Wstation | Identifier of the weather station where the reading was taken. |
| WRdaytime | WREADINGS.WRdaytime | Date and time of the weather reading. |
| Estdaytime | WREADINGS.Estdaytime | Whether the WREADINGS.WRdaytime is estimated.
TRUE if the date/time is estimated, FALSE if the
reading was taken at a known date and time. |
| WRperson | WREADINGS.WRperson | The OBSERVERS.Initials of the person who took the reading. |
| WRnotes | WREADINGS.WRnotes | Textual notes on the weather reading. |
| Tempmin | TEMPMINS.Tempmin | The minimum temperature reading, if any, since the last minimum temperature reading at the weather station. |
| Tempmin_Curated | TEMPMINS.Curated | Whether or not this minimum temperature has been curated. |
| Tempmax | TEMPMAXS.Tempmax | The maximum temperature reading, if any, since the last maximum temperature reading at the weather station. |
| Tempmax_Curated | TEMPMAXS.Curated | Whether or not this maximum temperature has been curated. |
| RGspan | RAINGAUGES.RGspan | The time elapsed since the rain gauge was last emptied. |
| EstRGspan | RAINGAUGES.EstRGspan | Weather or not the time elapsed since the rain
gauge was last emptied is an estimate. TRUE when
the elapsed time is based on one or more estimated
times, FALSE when the elapsed time is computed from
known endpoints. |
| Rain | RAINGAUGES.Rain | The amount of rain accumulation in millimeters. |
Inserting a row into MIN_MAXS inserts a row into
WREADINGS and rows into TEMPMINS, TEMPMAXS, and
RAINGAUGES as expected. Rows are only
inserted into TEMPMINS, TEMPMAXS, and RAINGAUGES
when the relevant columns are present and contain
non-NULL values.
Warning
Attempts to specify the WRid column on insert are
silently ignored. When inserting a new weather reading
the WRid column should be unspecified (the column
omitted or the data values specified as NULL). Babase
automatically computes a WRid and uses it appropriately in
the new rows.
Warning
The value of the RGspan and EstRGspan columns are ignored and
automatically computed values are used in their place.
It is best to omit these columns from the inserted data
(or specify them as NULL).
Warning
The PostgreSQL nextval()
function cannot be part of an INSERT
expression which assigns a value to this view's Wrid
column.
Deleting a row from MIN_MAXS deletes a row from WREADINGS and rows from TEMPMINS, TEMPMAXS, and RAINGAUGES as expected.
Contains one row for every row in WREADINGS. This view is the MIN_MAXS view sorted for ease of maintenance.
This view is less efficient than MIN_MAXS like view.
Figure 6.169. Query Defining the MIN_MAXS_SORTED View
SELECT wreadings.wrid AS wrid
, wreadings.wstation AS wstation
, wreadings.wrdaytime AS wrdaytime
, wreadings.estdaytime AS estdaytime
, wreadings.wrperson AS wrperson
, wreadings.wrnotes AS wrnotes
, tempmins.tempmin AS tempmin
, tempmins.curated AS tempmin_curated
, tempmaxs.tempmax AS tempmax
, tempmaxs.curated AS tempmax_curated
, raingauges.rgspan AS rgspan
, raingauges.estrgspan AS estrgspan
, raingauges.rain AS rain
FROM wreadings
LEFT OUTER JOIN tempmins
ON wreadings.wrid = tempmins.wrid
LEFT OUTER JOIN tempmaxs
ON wreadings.wrid = tempmaxs.wrid
LEFT OUTER JOIN raingauges
ON wreadings.wrid = raingauges.wrid
ORDER BY wreadings.wrdaytime, wreadings.wstation;;
Table 6.81. Columns in the MIN_MAXS_SORTED View
| Column | From | Description |
|---|---|---|
| WRid | WREADINGS.WRid | Identifier of the manual weather reading. |
| Wstation | WREADINGS.Wstation | Identifier of the weather station where the reading was taken. |
| WRdaytime | WREADINGS.WRdaytime | Date and time of the weather reading. |
| Estdaytime | WREADINGS.Estdaytime | Whether the WREADINGS.WRdaytime is estimated.
TRUE if the date/time is estimated, FALSE if the
reading was taken at a known date and time. |
| WRperson | WREADINGS.WRperson | The OBSERVERS.Initials of the person who took the reading. |
| WRnotes | WREADINGS.WRnotes | Textual notes on the weather reading. |
| Tempmin | TEMPMINS.Tempmin | The minimum temperature reading, if any, since the last minimum temperature reading at the weather station. |
| Tempmin_Curated | TEMPMINS.Curated | Whether or not this minimum temperature has been curated. |
| Tempmax | TEMPMAXS.Tempmax | The maximum temperature reading, if any, since the last maximum temperature reading at the weather station. |
| Tempmax_Curated | TEMPMAXS.Curated | Whether or not this maximum temperature has been curated. |
| RGspan | RAINGAUGES.RGspan | The time elapsed since the rain gauge was last emptied. |
| EstRGspan | RAINGAUGES.EstRGspan | Weather or not the time elapsed since the rain
gauge was last emptied is an estimate. TRUE when
the elapsed time is based on one or more estimated
times, FALSE when the elapsed time is computed from
known endpoints. |
| Rain | RAINGAUGES.Rain | The amount of rain accumulation in millimeters. |
The operations allowed are as described in the MIN_MAXS view.
In addition to the above views there are a number of views
which produce the group of a referenced individual as of a
pertinent date. These views are all named after the table from
which they are derived, with the addition of the suffixed
_GRP. They are nearly identical to the table
from which they derive, differing only by the addition of a
column named Grp.
The only operation allowed on these views is SELECT. INSERT, UPDATE, and DELETE are not allowed.
Figure 6.171. Query Defining the BIRTH_GRP View
SELECT biograph.*
, members.grp AS grp
FROM members, biograph
WHERE members.sname = biograph.sname
AND members.date = CAST(biograph.birth AS DATE);
Figure 6.173. Query Defining the ENTRYDATE_GRP View
SELECT biograph.*
, members.grp AS grp
FROM members, biograph
WHERE members.sname = biograph.sname
AND members.date = CAST(biograph.entrydate AS DATE);
Figure 6.175. Query Defining the STATDATE_GRP View
SELECT biograph.*
, members.grp AS grp
FROM members, biograph
WHERE members.sname = biograph.sname
AND members.date = CAST(biograph.statdate AS DATE);
Figure 6.177. Query Defining the CONSORTDATES_GRP View
SELECT consortdates.*
, members.grp AS grp
FROM members, consortdates
WHERE members.sname = consortdates.sname
AND members.date = CAST(consortdates.consorted AS DATE);
Figure 6.179. Query Defining the CYCGAPDAYS_GRP View
SELECT cycgapdays.*
, members.grp AS grp
FROM members, cycgapdays
WHERE members.sname = cycgapdays.sname
AND members.date = CAST(cycgapdays.date AS DATE);
Figure 6.181. Query Defining the CYCGAPS_GRP View
SELECT cycgaps.*
, members.grp AS grp
FROM members, cycgaps
WHERE members.sname = cycgaps.sname
AND members.date = CAST(cycgaps.date AS DATE);
Figure 6.183. Query Defining the CYCSTATS_GRP View
SELECT cycstats.*
, members.grp AS grp
FROM members, cycstats
WHERE members.sname = cycstats.sname
AND members.date = CAST(cycstats.date AS DATE);
Figure 6.185. Query Defining the DARTINGS_GRP View
SELECT dartings.*
, members.grp AS grp
FROM members, dartings
WHERE members.sname = dartings.sname
AND members.date = CAST(dartings.date AS DATE);
Figure 6.187. Query Defining the DISPERSEDATES_GRP View
SELECT dispersedates.*
, members.grp AS grp
FROM members, dispersedates
WHERE members.sname = dispersedates.sname
AND members.date = CAST(dispersedates.dispersed AS DATE);
Figure 6.189. Query Defining the MATUREDATES_GRP View
SELECT maturedates.*
, members.grp AS grp
FROM members, maturedates
WHERE members.sname = maturedates.sname
AND members.date = CAST(maturedates.matured AS DATE);
Figure 6.191. Query Defining the MDINTERVALS_GRP View
SELECT mdintervals.*
, members.grp AS grp
FROM members, mdintervals
WHERE members.sname = mdintervals.sname
AND members.date = CAST(mdintervals.date AS DATE);
Figure 6.193. Query Defining the MMINTERVALS_GRP View
SELECT mmintervals.*
, members.grp AS grp
FROM members, mmintervals
WHERE members.sname = mmintervals.sname
AND members.date = CAST(mmintervals.date AS DATE);
[244] Those columns that are joined in the view appear twice in the view ER diagrams, once in each of the two underlying tables. These columns appear only once in the view, so both the name of both columns in the view ER diagrams are followed by parenthesis containing the same text -- the name the column has in the view.
[245] Or attempts to update, as Babase may not allow these columns to be updated.
[246] Or attempts to update, as Babase may not allow these columns to be updated.
[247] Deletion is done on the DEMOG_CENSUS view in a fashion identical to the way it is done on the CENSUS_DEMOG view.
[248] Compared to the ordinal ranks in RANKS, which do not. E.g. a Rank of 2 might (or
might not) have a very different meaning when there are 20
total individuals, compared to another case when there are
2.
[249] Normally, Series would not be enough to determine the correct HPSId, but is okay in this case because TId and/or HSId are also required.
[250] It is technically _possible_ to allow this view to accept updates to the other columns, and thus allow updates to the WPRId, WPDId, and WPAId columns in WP_HEALUPDATES. However, unless the user is willing to address most or all of the columns in this view in their update command, the validation needing to be written for these updates is prohibitively lengthy. The yield in utility does not seem worth the time investment to write such a thing, hence these kinds of updates are prohibited.
[251] It is possible to improve the algorithm used to
discern valid observer codes. This would reduce the need
for manual intervention on the part of the data manager at
the cost of increased complexity in the code. Given the
extreme UNlikelihood that there will ever be an observer
whose initials include a
"/", this improvement
seems unnecessary.
[252] There is little use in attempting to update CYCLES because updates to the the Seq and Series columns are silently ignored and changing Sname is not allowed.
[253] Or attempts to update, as Babase may not allow these columns to be updated.
[254] There is little use in attempting to update CYCLES because updates to the the Seq and Series columns are silently ignored and changing Sname is not allowed.
[255] Or attempts to update, as Babase may not allow these columns to be updated.
[256] This is implicit, because if she also has no data in REPRO_NOTES, then there won't be a row in the view at all.
[257] Babase should contain one and exactly one actor and one and exactly one actee for every interaction. The ACTOR_ACTEES view does left outer joins of the INTERACT_DATA table with the PARTS table so that invalid data can still be maintained in the event the stated relationships do not exist. To ignore invalid data, e.g. for purposes of analysis, write a query that does regular joins instead of the left outer joins used by the view.
[258] This restriction is leftover from implementation limitations in older versions of PostgreSQL and could now be removed if so desired.
[259] Babase should contain one and exactly one actor and one and exactly one actee for every interaction. The MPI_EVENTS view does left outer joins of the MPI_DATA table with the MPI_PARTS table so that invalid data can be observed in the event the stated relationships do not exist. To ignore invalid data, e.g. for purposes of analysis, write a query that does regular joins instead of the left outer joins used by the view.
[260] 3 * 2 = 6 (Doh!)
[261] Typically the coalition id numbers increase sequentially, but the program does not require this.
[262] It is not usually a good idea to have the upload program perform such simple transformations because it eliminates any flexibility in the codes chosen for use. However in this case there is advantage in having the uploaded files more closely resemble the field data. Right?
[263] Other integrity checks are left to the database to perform.
[264] This is expected to be the group that is sampled, but you never know.
[265] Note that when is no additional anesthetic administered there are no related ANESTHS rows and hence no row in this view for the given darting.
[266] Note that when no body temperature measurements are taken there are no related BODYTEMPS rows and hence no row in this view for the given darting.
[267] Note that when no chest circumference measurements are taken there are no related CHESTS rows and hence no row in this view for the given darting.
[268] Note that when no crown-to-rump measurements are taken there are no related CROWNRUMPS rows and hence no row in this view for the given darting.
[269] Note that even when no collected samples are recorded for a particular darting in DART_SAMPLES, there will still be a row for that darting in this view.
[270] Note that when no tooth observations are recorded there are no related TEETH rows and hence no row in this view for the given darting.
[271] Note that when no tooth observations are recorded there are no related TEETH rows and hence no row in this view for the given darting.
[272] Column names cannot begin with a digit so the letter
s, for “site”, is used to
preface the name of each column.
[273] Note that when no humerus length measurements are taken there are no related HUMERUSES rows and hence no row in this view for the given darting.
[274] Note that when no PCV measurements are taken there are no related PCVS rows and hence no row in this view for the given darting.
[275] Note that when no testes length or width measurements are taken there are no related TESTES_ARC rows and hence no row in this view for the given darting.
[276] Note that when no testes length or width measurements are taken there are no related TESTES_DIAM rows and hence no row in this view for the given darting.
[277] Note that when no ulna length measurements are taken there are no related ULNAS rows and hence no row in this view for the given darting.
[278] Note that when no vaginal pH measurements are taken there are no related VAGINAL_PHS rows and hence no row in this view for the given darting.
[279] Allowing updates via the the LocalId columns is certainly doable, but requires an uncomfortable amount of "magic". At the time of this writing it doesn't seem like a huge burden to only allow direct updates to NAId.
[280] Or, in a single transaction.
[281] Because sleeping grove information is entered into the
GPS units as 2 separate waypoints, which spreads the
sleeping grove information over two lines of uploaded data,
the A and
D codes are always
used when the SWERB_LOC_DATA row is
created by the SWERB_UPLOAD view. The
SWERB_LOC_DATA.ADcode value is then updated to
the correct value when the 2nd SWERB waypoint is
processed.
The alternative to this temporary use of the
A and
D codes is to allow
the SWERB_LOC_DATA.ADcode to be NULL until the
transaction is committed, and to defer
related checks until transaction commit.
[282] This is due to the 10 character data entry limit in the GPS units. The entry of an S character when recording drinking events would cause the 10 character limit to be exceeded (when the waterhole codes are 4 characters, as they often are) so the SWERB_UPLOAD view uses this method to guess whether a subgroup was observed.
[283] Unusual to those familiar with the data collection protocol. We don't mention things that might be surprising to the casual observer.
[284] This is not ideal. Or rather, the approach is sound but the practice deficient. The Amboseli Baboon Research Project Monitoring Guide's description of the waypoint text entered by the operator could use some work.
[285] The only time this is an issue is when a team is observing more than one group at one time. In this case manual intervention on the part of the data manager is also required to avoid double counting observer effort. See the Is_Effort column.
[286] The SWERB_UPLOAD view does not actively test for these conditions, it assumes that they exist. In the normal course of events it is unlikely, but possible, to insert invalid data into SWERB when these conditions are violated.
[287] It is possible to improve the algorithm used to better discern valid observer codes. This would reduce the need for manual intervention on the part of the data manager at the cost of increased complexity in the code.
[288] In this case, it is assumed but not required that the data manager will also make good use of the Timeest and Source columns.
[289] Or at least this is the ideal. In actual practice it is difficult for the data managers to know when a group has been observed by more than one team on any given day.
[290] Bouts of observation of zero-length, although the system does not require this. Note that the Is_Effort column may be of interest in these cases.
[291] Note that the rule which requires a strict time-wise ordering of the uploaded rows does not apply to the begin and end rows marked as secondary ascents and descents. This allows the creation of secondary ascent and descent bouts which are of non-zero length, should such need to exist.
[292] Because the MIN_MAXS view always returns a row regardless of whether data exists in TEMPMINS, TEMPMAXS, and RAINGAUGES the view may sometimes be less useful than, say, a query which returns only those rows where there is both a minimum and a maximum temperature reading. In other words, as usual, it's always prudent to know what you're doing when querying Babase.
Table of Contents
The prototypical way to import data into Babase is in bulk, via a plain text file having columns delimited by the tab character. These are easily produced by almost any spreadsheet program; it is expected that most data imported into Babase will be typed into a spreadsheet and then exported to tab-delimited text for upload.
Most data are uploaded into Babase via the Upload program, most often directly into Views. The phpPgAdmin program's import function can also be used, although it does not allow import into views. Data may also be entered row by row directly into the database, either via the phpPgAdmin web interface, or by entering SQL into either phpPgAdmin or any other PostgreSQL front-end.[293]
Babase contains a number of bespoke programs , including some dedicated Data Maintenance Programs and Views. Some few of these programs are utility in nature: a program to logout, a program to automate the steps involved in the creation of a new database user, and so forth. Most of the data entered into Babase is collected in tabular, row-and-column, format suitable for entry into a relational database. As mentioned previously, this data can be imported directly. The purpose of most of the bespoke Babase programs is to transform data files, as part of the data upload process, from the formats easily collected in the field into a tabular format.
Most errors in computer data entry can be caught by the wwwdiff program. This program compares files. Typos are detected by entering the data twice, preferably by 2 different people, and comparing the result. Errors made in the field are more likely to be detected by manual checks of the data, or by the data validation built into Babase.
The system will automatically generate id columns whenever a new row is inserted and an id column is not supplied. When an id value is supplied the system does not check to see that it is indeed the next id in sequence, nor does it update the sequence number to be automatically supplied the next time a row is inserted without an id column. Should an id value be manually supplied, it may be necessary to update the internal id counter so that future system generated ids will not conflict with an id already used.
See the Postgresql documentation section on sequence functions for reference material on the requisite PostgreSQL functions.
Tip
Don't supply id values manually unless you know what your doing.
Automatically generated ids are not guaranteed to be contiguous.
[293] The psql program runs from the Unix prompt. It allows you to type SQL queries interactively and see the results. (phpPgAdmin submits one or more SQL statements to the database to be executed and then disconnects after getting the results back.) Psql has “meta-commands” that do things like report on database structure, and it facilities writing scripts to automate a wide variety of tasks. Examples of such scripts can be found in the Babase source code.
Other possible front-ends are discussed in another footnote.
Table of Contents
These are the programs and views that are used in the entry and maintenance of the Babase Master tables. Their use is fully documented in the procedure manual. The summary written here provides a statement of purpose and a mention of all updated data. The operation and behavior of the programs and views supports the table and program characteristics documented in this manual. For more information on the actual capabilities of the programs and views see the documentation in the headings of the programs' source code and the source code of the views' triggers .
The programs and views are designed to upload data in “batch” -- each run of a program uploads a single file containing multiple lines of data, each of which is then inserted into the database as a row of data in one (or more[294]) database tables.
The views presented in this section are not intended to be useful when querying. They exist to provide an upload mechanism for updating tables, and are views rather than tables to simplify overall system maintenance.
Most of the upload programs, the exception being the Psionload program, take as input a file of data arranged in tabular format. The file is expected to contain plain text, with rows on separate lines and columns separated by a single tab character. This data structure can be produced by exporting data from a spreadsheet as tab delimited text.
The programs and views upload the data into the database in an all-or-nothing fashion. Ether all the data in a uploaded file is inserted into the database or, should any error occur, none of it is. After an error the processing of the uploaded file continues so as to catch additional errors. However the input line containing the erroneous data is ignored, so the “trial”[295] insertion into the database of the subsequent lines in the uploaded file may result in spurious errors due to the “missing” data. It is left to the operator to distinguish the real errors from the false positives.
Tip
When in doubt simply correct the errors that are clearly problems, notably the first error reported, and re-run the program or re-upload into the view.
Tip
For reasons of security most browsers will remove pathnames from forms. Should a program which imports data into Babase from a file find an error in the data, rather than re-enter the pathname of the file to be uploaded simply press the browser's "reload" button. This (usually, depending on the browser) redoes the upload using the previously entered file name -- but with the new, now-corrected, data content.
Each time any of the Babase web programs successfully uploads a file into a database Babase remembers the name of the file and the database. None of the Babase programs will allow the same user to re-upload a file of the same name into the same database, until either a file with a different name is loaded into that database or until the user logs out and back in.
For more information on whether data is required to present, as well as other required characteristics of the data values, see the documentation of the specific column into which the data is stored.
The SWERB_UPLOAD view takes the place of an upload program. The Upload program can be used to insert data into this view and thence into the various SWERB Data tables.
The upcen program updates the CENSUS table. It is accessed over the web and can be found on the Babase Web site.
The upcen program updates the CENSUS table on a group-by-group basis. A single run of upcen can update CENSUS with multiple days of data on multiple individuals, but all the data must be for a single group.
Rows inserted into the CENSUS table by
upcen have a CENSUS.Cen value of TRUE.
Should a data validation error occur during the execution of upcen the CENSUS table will not be updated at all. Upcen runs in an all or nothing fashion, either all of the data supplied to it is entered into the database or none is.
Upcen takes a single file of census data arranged in tabular format, a format very similar to the data sheets filled out in the field. The file is expected to contain plain text, with rows on separate lines and columns separated by a single tab character. This data structure can be produced by exporting data from a spreadsheet as tab delimited text.
The layout of the data in the file is as follows:
- First Cell
The first cell of data, the one in the first column of the first row, must contain the GROUPS.Gid code for the group.
- First Row
The remainder of the first row of data, the entire first row excepting the first cell, must contain the dates on which the census was taken.
Tip
In order to avoid confusion between European and American date styles, and other sorts of foolery with Excel dates, it may be a good idea to have the spreadsheet format this data as text rather than as dates.
- First Column
The remainder of the first column of data, the entire first column excepting the first cell, must contain the Snames of the censused individuals.
- Census Data
The remainder of the table, everything excepting the first row and first column, contains the census data. Each cell represents the census taken of an individual, who's Sname appears in the first column, on a census date, the date appearing in the first row. Each cell can contain one of three possible values:
NWhen
N(upper case letter N, meaning “No data”) appears in the cell there was no census of the given individual on the given date. Upcen does nothing to the CENSUS table.- No Data
When no data appears in the cell the individual was censused present. The CENSUS table is updated with a Status code of
Cfor that individual/group/day.0When a
0(digit zero) appears in the cell then individual was censused absent. The CENSUS table is updated with a Status code ofAfor that individual/group/day.
The MPI_UPLOAD view takes the place of an upload program. The Upload program can be used to insert data into this view and thence into the various tables related to MPIS.
The updart program uploads darting data into Babase.
As with the other data entry programs all data in the uploaded file is recorded in the database in an all or nothing fashion; the database is unchanged if any errors occur.
The updart program accepts a variety of data formats depending on the type of darting data uploaded. The format of the uploaded data is determined by the menu selection used to invoke the updart program.
Caution
For any given darting logistic data must be uploaded first. The remaining data can be uploaded in any order.
The updart program will not overwrite data on the DARTINGS table. The textual note columns on DARTINGS must be NULL before being replaced with
a value. In some cases this will help prevent the uploading
of duplicate data.[296] Updart also reports an error when successive
lines in the uploaded file have identical
sname and dartdate
values.[297]
Caution
Because much of the darting data can involve collection of multiple sets of repeated data per darting there are few checks which prevent duplicate data.
By way of example, there are no restrictions which require that all the data which pertain to a given darting be recorded in contiguous rows so repetition of a darting in a later part of an uploaded file is not detected. Care must be taken not to upload the same data twice.
Each line in the uploaded file corresponds to the darting of a single individual. The uploaded file may contain leading or trailing empty lines. No data must be indicated by an empty cell.
The uploaded file must begin with a line of column headings with the names given in each section below in the order given in the sections below. The column headings are validated but otherwise unused, with the exception of the “numbered” columns that appear in sets as described in the next paragraph. The checking of column names is to assist in the detection of data entry errors. The content of each column is as described.
The “numbered” columns, such as the
columns labeled extra_anesthN,
extra_anesth_timeN, and
extra_anesth_amtN, must be supplied in
matching sets. The N in the column name
is presented here as a placeholder and the counting numbers
1, 2,
3, etc., must be substituted in actual
use. The set of columns may be repeated as many times as
needed, or not used at all, the restriction being that the
first occurrence must use column names ending in the number
1 with successive repetitions
incrementing the “column number” by one. When
the uploaded data has more sets of columns than are needed
for a given line, a given darting, the unneeded columns are
to be left empty.
Every data format accepted by the updart program begins with the following columns in the order written here:
nameThe name of the darted individual. The given value is compared in a case-insensitive fashion with BIOGRAPH.Name but is otherwise unused.
This column must contain a value.
This data is not recorded in the database but is checked for validity to assist in detection of data entry errors.
snameThe BIOGRAPH.Sname of the darted individual.
This column must contain a value.
When supplied with other darting logistic data this data is stored in the DARTINGS.Sname column. Otherwise it is used together with the
dartdatecolumn to identify the related DARTINGS row.sexThe sex of the darted individual, either
Mfor male orFfor female.This column must contain a value.
This data is not recorded in the database but is checked against BIOGRAPH.Sex to assist in detection of data entry errors. The data in this column is not otherwise used.
dartdateThe date the individual was darted. When supplied with other darting logistic data this data is stored in the DARTINGS.Date database column. Otherwise it is used together with the
snamecolumn to identify the related DARTINGS row.
Logistic data is uploaded into the DARTINGS, and ANESTHS tables.
In addition to the initial columns common to all the updart upload formats the logistic data format contains the following columns:
darttimeThe time the individual was darted. This data is stored in the DARTINGS.Darttime database column.
downtimeThe time the individual succumbed to the anesthetic, the DARTINGS.Downtime value.
pickuptimeThe time the individual was picked up by the darting team. This value is stored in the DARTINGS.Pickuptime column.
dartdrugThe ANESTHS.Drug of the anesthetic delivered by dart. This value is stored in the DARTINGS.Drug column.
extra_anesthNThe type of extra anesthetic administered, an DRUGS.Drug value. This value is stored in the ANESTHS.Drug column.
extra_anesth_timeNThe time extra anesthetic was administered. This value is stored in the ANESTHS.Antime column.
extra_anesth_amtNThe amount of extra anesthetic administered. This value is stored in the ANESTHS.Anamount column.
other_notesTextual notes related to darting logistics. This value is stored in the DARTINGS.Logisticnotes column.
commentsGeneral comments on the darting. This value is stored in the DARTINGS.Dartcomments column.
Morphology data is uploaded into the DARTINGS, CROWNRUMPS, CHESTS, ULNAS, and HUMERUSES tables.
In addition to the initial columns common to all the updart upload formats the morphology data format contains the following columns:
bodymassThe individual's mass. This data is stored in the DARTINGS.Mass database column.
crownrumpNThe crownrump measurement. This data is stored in the CROWNRUMPS.CRlength database column.
crobserverNThe observer who took the crownrump measurement. This data is stored in the CROWNRUMPS.CRobserver database column.
chestcircumNThe chest circumference measurement. This data is stored in the CHESTS.Chcircum database column.
unadj_chestcircumNThe unadjusted chest circumference measurement. This data is stored in the CHESTS.Chunadjusted database column.
chobserverNThe observer who took the chest circumference measurement. This data is stored in the CHESTS.Chobserver database column.
ulnaNThe ulna measurement. This data is stored in the ULNAS.Ullength database column.
unadj_ulnaNThe unadjusted ulna measurement. This data is stored in the ULNAS.Ulunadjusted database column.
ulobserverNThe observer who took the ulna measurement. This data is stored in the ULNAS.Ulobserver database column.
humerusNThe humerus measurement. This data is stored in the HUMERUSES.Hulength database column.
unadj_humerusNThe unadjusted humerus measurement. This data is stored in the HUMERUSES.Huunadjusted database column.
huobserverNThe observer who took the humerus measurement. This data is stored in the HUMERUSES.Huobserver database column.
crnotesNotes on the crownrump measurements This data is stored in the DARTINGS.CRnotes database column.
chnotesNotes on the chest circumference measurements This data is stored in the DARTINGS.Chnotes database column.
ulnotesNotes on the ulna measurements This data is stored in the DARTINGS.Ulnotes database column.
hunotesNotes on the humerus measurements This data is stored in the DARTINGS.Hunotes database column.
Physiology data is uploaded into the DARTINGS, DPHYS, PCVS, and BODYTEMPS tables.
In addition to the initial columns common to all the updart upload formats the physiology data format contains the following columns:
hematocritNThe individual's PVC. This data is stored in the PCVS.PCV database column.
bodytempNThe individual's body temperature. This data is stored in the BODYTEMPS.Btemp database column.
bodytemptimeNTime the individual's body temperature was taken. This data is stored in the BODYTEMPS.Bttime database column.
pulseIndividual's pulse. This data is stored in the DPHYS.Pulse database column.
respirationIndividual's respiration. This data is stored in the DPHYS.Respiration database column.
r_inguinal_lymphState of the individual's right inguinal lymph node. This data is stored in the DPHYS.Ringnode database column.
l_inguinal_lymphState of the individual's left inguinal lymph node. This data is stored in the DPHYS.Lingnode database column.
r_axillary_lymphState of the individual's right axillary lymph node. This data is stored in the DPHYS.Raxnode database column.
l_axillary_lymphState of the individual's left axillary lymph node. This data is stored in the DPHYS.Laxnode database column.
r_submandibular_lymphState of the individual's right submandibular lymph node. This data is stored in the DPHYS.Rsubmandnode database column.
l_submandibular_lymphState of the individual's left submandibular lymph node. This data is stored in the DPHYS.Lsubmandnode database column.
other_notes_measuresNotes on physiological features. This data is stored in the DARTINGS.Dphysnotes database column.
pcvnotesNotes on PVC measurements. This data is stored in the DARTINGS.PCVnotes database column.
btempnotesNotes on body temperature measurements. This data is stored in the DARTINGS.Bodytempnotes database column.
This program is no longer functional. It was used to add data to the DSAMPLES table, which has been replaced by the DART_SAMPLES table and DSAMPLES view.
Physical sample related data is uploaded into the DARTINGS and DSAMPLES tables.
In addition to the initial columns common to all the updart upload formats the physical sample data format contains the following columns:
(In progress, to be added later)
Data related to teeth is uploaded into the DARTINGS and TEETH tables.
In addition to the
initial columns common to all the updart upload
formats the teeth data format contains the following
columns. Most of these columns are “special”
in that the column name is used to designate a related TOOTHCODES row, indicating the position of the
tooth within the mouth. The text written into the upload
file's column names shown here as
TOOTHCODE must be replaced with the
actual tooth code. Data related to each tooth code is
presented as a set comprising the tooth's state (TSTATES) and the tooth's condition (TCONDITIONS).
Note
Unlike the “numbered” column headers used with other sorts of repeating data all of the TEETH related columns need not be present. Their order is also not significant. However all of the columns pertaining to a particular tooth code must be adjacent.
TOOTHCODE_tstateThe state of the tooth. (E.g. present, erupting, missing, etc.) This data is stored in the TEETH.Tstate database column.
TOOTHCODE_tconditionThe condition of the tooth. (E.g. healthy, decayed, etc.) This data is stored in the TEETH.Tstate database column.
notesGeneral notes on the teeth. This data is stored in the DARTINGS.Teethnotes database column.
caninenotesGeneral notes on the canines. This data is stored in the DARTINGS.Caninenotes database column.
Testes related data is uploaded into the DARTINGS, TESTES_ARC, and TESTES_DIAM tables.
Caution
The determination of left or right testicle is not made based on the name of the column but by the value of the Testside or Testside column.
Note
The left and right side measurements are separate and distinct “numbered column” set. This means there need not be as many left as right side measurements.[298]
In addition to the initial columns common to all the updart upload formats the testes data format contains the following columns:
ltesteslengthNThe length of the (left) testicle. This data is stored in the TESTES_ARC.Testlength and TESTES_DIAM.Testlength database columns.
ltesteswidthNThe width of the (left) testicle. This data is stored in the TESTES_ARC.Testwidth and TESTES_DIAM.Testwidth database columns.
ltestessideNIndication of left or right testicle. It is presumed but not required that a value of
Lbe supplied indicating the length and width are of the left testicle. This data is stored in the TESTES_ARC.Testside and TESTES_DIAM.Testside database columns.rtesteslengthNThe length of the (right) testicle. This data is stored in the TESTES_ARC.Testlength and TESTES_DIAM.Testlength database columns.
rtesteswidthNThe width of the (right) testicle. This data is stored in the TESTES_ARC.Testwidth and TESTES_DIAM.Testwidth database columns.
rtestessideNIndication of left or right testicle. It is presumed but not required that a value of
Rbe supplied indicating the length and width are of the right testicle. This data is stored in the TESTES_ARC.Testside and TESTES_DIAM.Testside database columns.other_notes_measuresNotes regarding testicle measurements. This data is stored in the DARTINGS.Testesnotes database column.
The uptick program uploads into Babase data on parasite infestation collected during dartings. For any given darting it must be run after the darting logistic data is uploaded.
Each line of the uploaded file corresponds to a parasite count of a particular body part taken during a specific darting -- corresponds to a row in the TICKS table.
As with the other data entry programs all data in the uploaded file is recorded in the database in an all or nothing fashion; the database is unchanged if any errors occur.
The uptick program will not overwrite data in the DARTINGS.Ticknotes
column. This column must be NULL before being replaced with
a value. In some cases this will help prevent the uploading
of duplicate data.
Caution
Because much of the darting data can involve collection of multiple sets of repeated data per darting there are few checks which prevent duplicate data.
By way of example, there are no restrictions which require that all the data which pertain to a given darting be recorded in contiguous rows so repetition of a darting in a later part of an uploaded file is not detected. Care must be taken not to upload the same data twice.
The uploaded file may contain leading or trailing empty lines. No data must be indicated by an empty cell.
The uploaded file must begin with a line of column headings with the names given below in the order given below. The column headings are validated but otherwise unused. This is to assist in the detection of data entry errors. The content of each column is as described.
Aside from the line containing the column headings the
uploaded rows can be ordered in any fashion. There is no
requirement that the rows pertaining to a single darting be
contiguous. However, because DARTINGS.Ticknotes cannot be overwritten only one
uploaded row per darting may have a non-empty
other_notes_measures cell.
nameAs described in Updart.
snameAs described in Updart.
sexAs described in Updart.
dartdateAs described in Updart.
pcountThe number of parasites found on the designated body part. This data is stored in the TICKS.Tickcount database column.
bodypartThe body part examined for parasites. This data is stored in the TICKS.Bodypart database column.
pkindThe kind of parasite counted. This data is stored in the TICKS.Tickkind database column.
pstatusThe classification of the count itself. This data is stored in the TICKS.Tickstatus database column.
pnotesNotes on the counting of the parasites. This data is stored in the TICKS.Tickbpnotes database column.
other_notes_measuresGeneral notes on the counting of ticks and other parasites. This data is stored in the DARTINGS.Ticknotes database column.
Because the
other_notes_measurescolumn is stored on DARTINGS there can only be one per darting. To ensure this only the first row for any given darting may contain a value forother_notes_measures, the remaining cells for the darting must be empty.
Psionload transfers the output of the Psion palmtop computers' focal point sampling data into Babase.
The Psionload program only knows how to load data with
the semantics of the data structure described by DATA_STRUCTURES.Data_Structure value
1. This format is
documented on the Psion Data Format
page of the Babase
Wiki.
Note that the time recorded in a Psion ad-lib row is stored in both the Start and Stop columns of the INTERACT_DATA table.
This program makes a lot of assumptions about the contents of the STYPES, ACTIVITIES, POSTURES, and NCODES tables. It was written when those tables were laden with special values[299] to support two and only two sample types: samples on adult females and samples on juveniles of any sex. In the production database, these tables are appropriately configured and this program should function normally. But if installed in a new, "clean" database, this program will certainly NOT work.
Warning
The psionload program assumes that every program[300] that uses a setupfile[301] produces an output file having identical structure and semantics. If this assumption is violated then the data will either not load, or worse yet, will load in an incorrect fashion.
Any changes in the form or semantics of the data collected with the Psions must be indicated in the Psion data by way of a change in the Psion setup id string and the DATA_STRUCTURES row referenced thereby. If the setup id string does not reflect changes in the Psion data then the data will either not load, or worse yet, will load in an incorrect fashion.
Upload uploads data into any table or view. Its primary purpose is to upload data into views; at the time of this writing PostgreSQL and its various front ends are unable to import data into views.
Tip
The name of the table may be qualified with a schema name to upload data into tables or views that are not in the babase schema.
There are 2 ways to upload NULL data values. The
easiest is to omit the column. Columns without some other
default value will be given NULL values. The second is to
check the checkbox labeled "Upload NULL Values"
and supply a input value for NULL. Data values that match
the given NULL representation will then be given a NULL
value in the database.
The default NULL representation is the empty string,
no data at all. When this representation is used data that
are omitted in the input file becomes NULL when uploaded
into the database.
Caution
A space (or multiple spaces) may be chosen as the
NULL representation. This can be difficult to discern
while operating the program.
Data to be uploaded must be in tab delimited format. The first line of the input file must contain the column names, each separated by a tab. The remaining lines of the file contain the data to be uploaded. Each line is a row of data, each column is separated from its neighbor with a tab character.
A line need not contain as many tab separated data elements as there are column names given in the first line. All unspecified data elements will be given a blank value, the empty string, just as if the tabs occurred but no data were specified.[302]
A line must not have more tab separated data elements than there are column names given in the first line.
This section describes programs and functions available for general use. These functions are in addition to those supplied as part of the PostgreSQL system. Typically, one would use one of these programs as part of a special process not part of the regular Babase system. One would use one of these functions in a SQL - SELECT statement, a query, a report, or perhaps in a special purpose program, or a new Babase system program you might want to write. For more detailed information on the operation of these programs and functions see the documentation written into the program header of the program source code.
Documentation on the use of these programs can be found in the Protocol for Data Management: Amboseli Baboon Project and in this document. This document also contains the coding standards and design philosophy of the system, which should be followed by anyone modifying or adding programs to this directory.
Note
There are a large number of procedures and functions
which are part of Babase but are not listed in the tables below.
The “unlisted” functions either begin with the
underscore (_) character, or end in
“_func”.[303]
Only those functions in the table below are expected to be used by the typical end-user.
Table 8.1. The Babase SQL Functions
| Name | Description |
|---|---|
|
convert a date value to the first day of the month | |
|
return a remainder from a timestamp | |
|
convert time to seconds past midnight | |
|
convert from seconds past midnight to a time | |
|
convert a date or timestamp to a julian date | |
|
convert a julian date to a regular date | |
|
compute the hydrological year a given date falls within | |
|
compute the season a given date falls within | |
|
produce a PostGIS geometry point in the WGS 1984 UTM Zone 37South coordinate system from X and Y coordinates | |
|
produce a PostGIS geometry point in the WGS 1984 UTM Zone 37South coordinate system from longitude and latitude coordinates | |
|
Convert a "raw" hormone concentration into a corrected concentration, using the provided mathematical expression. | |
|
convert a quantity from one given unit to another. | |
|
convert an interval into a string with format "HH:MM:SS". E.g. 1 day 02:03:04 becomes 26:03:04. |
Name
rnkdate — convert a date value to the first day of the month
Synopsis
date
rnkdate
( | dateval); |
date
dateval
;
timestamp
rnkdate
( | dateval); |
timestamp
dateval
;Description
A function which returns a date that is the first day of the month of a given date value.
This function is similar to the
date_trunc('month', date) function, but takes
care of data type conversions and handles time zones. The
postgresql date_trunc() function expects a
timestamp or timestamptz data
type as input. This can lead to confusion involving time
zones.
Warning
Always use the rnkdate() function
when comparing a date with a RANKS.Rnkdate value.
Unless care is taken, use of the Postgresql
date_trunc() function can lead to
mis-matches due to time zone complications.
While it may well be possible to use the
date_trunc() function in place of the
rnkdate() function; the possibility of
complications due to time zone issues has not been
thoroughly investigated. Better safe than sorry.
Name
date_mod — return a “remainder” from a timestamp
Synopsis
interval
date_mod
( | period, | |
daytime); |
text
period
;
timestamp
daytime
;
interval
date_mod
( | period, | |
daytime); |
text
period
;
date
daytime
;
interval
date_mod
( | period, | |
daytime); |
text
period
;
interval
daytime
;
interval
date_mod
( | period, | |
daytime); |
text
period
;
time
daytime
;Input
period
A string indicating a unit of time. The values
allowed are the same as those allowed for the
field parameter of the PostgreSQL
date_trunc
function. At the time of this writing these
are:
microsecondsmillisecondssecondminutehourdayweekmonthquarteryeardecadecenturymillennium
Description
A function which returns the interval
“remaining” from the given
daytime value after the last whole
period. I.e. date_mod('hour',
'1941-12-7 07:45:00'::timestamp) returns the
interval following the last hour, 45:00
minutes.
date_mod() operates on timestamp
and date values in a fashion conceptually similar to the
numeric modulo function, which returns the remainder after
division.
Name
spm — convert time to seconds past midnight
Synopsis
double precision
spm
( | tvalue); |
time
tvalue
;
double precision
spm
( | tvalue); |
interval
tvalue
;
double precision
spm
( | tvalue); |
timestamp
tvalue
;
int
spm
( | tvalue); |
time(0)
tvalue
;Description
A function which returns the number of seconds past midnight. When given a timestamp rather than a time it returns the number of seconds past midnight in the time portion of the timestamp. When given an interval rather than a time it returns the number of seconds in the interval, ignoring any whole days in the interval.
This function is useful for the analysis of intervals.
Name
spm_to — convert from seconds past midnight to a time
Synopsis
time
spm_to
( | secs); |
double precision
secs
;
time(0)
spm_to
( | secs); |
int
secs
;Name
julian — convert a date or
timestamp to a julian date
Synopsis
INT
julian
( | date); |
DATE
date
;
INT
julian
( | date); |
TIMESTAMP
date
;Description
You supply this function with a date
(or a timestamp) and it returns the integer
that represents the given date as the number of days since
a particular reference date. This number is known as the
Julian date
representation of the given date. (Day number 2,361,222 is
September 14, 1752.) Legal values for the date are between
September 14, 1752 and December 31, 9999,
inclusive.
Name
julian_to — convert a julian date to a regular date
Synopsis
DATE
julian_to
( | julian_date); |
INT
julian_date
;Description
This function reverses the julian() function. You supply this function with a julian date and it returns a regular date.
Name
hydroyear — compute the hydrological year a given date falls within
Synopsis
INT
hydroyear
( | date); |
DATE
date
;
INT
hydroyear
( | timestamp); |
TIMESTAMP
timestamp
;
INT
hydroyear
( | textualdate); |
TEXT
textualdate
;Description
Return the hydrological year as an
integer given a date or
timestamp. The hydrological year begins
November 1 and ends October 31. The number associated
with a hydrological year is the calendar year in which the
majority of the hydrological year falls, the calendar year
of the October 31 at the end of the hydrological
year.
Name
season — compute the season a given date falls within
Synopsis
CHAR(1)
season
( | date); |
DATE
date
;
CHAR(1)
season
( | timestamp); |
TIMESTAMP
timestamp
;
CHAR(1)
season
( | textualdate); |
TEXT
textualdate
;Name
bb_makepoint — produce a PostGIS geometry point in the WGS 1984 UTM Zone 37South coordinate system from X and Y coordinates
Synopsis
geometry
bb_makepoint
( | X, | |
Y); |
double precision
X
;
double precision
Y
;Input
X
The X coordinate of the point in the WGS
1984 UTM Zone 37South
coordinate system. A double precision
number, or any other data type that can be interpreted
as a number and converted to double precision.
Y
The Y coordinate of the point in the WGS
1984 UTM Zone 37South
coordinate system. A double precision
number, or any other data type that can be interpreted
as a number and converted to double precision.
Description
Return a PostGIS geometry point in the WGS 1984 UTM Zone 37South coordinate system, the geolocation coordinate system used within Babase. Such points can be used in geospatial analysis.
This is a convenience function that is shorthand for
the PostGIS expression: ST_SetSRID(ST_MakePoint(x,
y), 32737). However, unlike
ST_MakePoint(), bb_makepoint requires that either both
x and y be NULL or
neither be NULL.
Name
bb_makepoint_longlat — produce a PostGIS geometry point in the WGS 1984 UTM Zone 37South coordinate system from longitude and latitude coordinates
Synopsis
geometry
bb_makepoint_longlat
( | Long, | |
Lat); |
double precision
Long
;
double precision
Lat
;Input
Long
The Longitude of the point in the WGS
1984 2D CRS,
in decimal degrees. A double precision
number, or any other data type that can be interpreted
as a number and converted to double precision.
Lat
The Latitude of the point in the WGS
1984 2D CRS,
in decimal degrees. A double precision
number, or any other data type that can be interpreted
as a number and converted to double precision.
Description
Return a PostGIS geometry point in the WGS 1984 UTM Zone 37South coordinate system, the geolocation coordinate system used within Babase. Returning a PostGIS geometry point in the WGS 1984 2D CRS — the system used in the provided coordinates — would not be especially helpful, because no tables in Babase use that system.
This is a convenience function that is shorthand for
the PostGIS expression:
ST_TRANSFORM(ST_SetSRID(ST_MakePoint(long, lat),
4326), 32737). However,
unlike ST_MakePoint(), bb_makepoint_longlat() requires
that either both long and
lat be NULL or neither be
NULL.
Name
corrected_hormone — Convert a "raw" hormone concentration into a corrected concentration, using the provided mathematical expression.
Synopsis
NUMERIC(8,2)
corrected_hormone
( | raw_conc, | |
correction); |
NUMERIC(8,2)
raw_conc
;
text
correction
;Input
raw_conc
The concentration in need of correction, e.g. a
HORMONE_RESULT_DATA.Raw_ng_g value. A
NUMERIC(8,2) number, or any other
data type that can be interpreted as a number and
converted to NUMERIC(8,2).
correction
A text string containing a mathematical expression
that indicates the arithmetic needed to correct the raw
concentration. Usually this will be a HORMONE_KITS.Correction value. When
referring to the "raw" value, the string
%s must be used[304]. Some example expressions are provided
below.
| Correction | Interpretation |
|---|---|
%s | Use the raw value, no correction needed |
%s /
100 | Divide the raw value by 100 |
(2 * %s) +
50 | Multiply the raw value by 2, then add 50 |
It is assumed that this mathematical correction is
based on a linear relationship, so %s
cannot appear more than once in the "correction".
Description
Returns a number, the corrected concentration. If
either or both of the parameters is NULL, returns
NULL.
This function is used by the ESTROGENS, GLUCOCORTICOIDS, HORMONE_RESULTS, PROGESTERONES, TESTOSTERONES, and THYROID_HORMONES views to generate their respective Corrected_ng_g columns. It may also be useful to users who want to try a different correction factor that is not recorded in HORMONE_KITS.
[304] Yes, %s is ugly, but this was
not chosen arbitrarily. It has a very specific meaning
for the PostgreSQL FORMAT() function, which is used in
this function to convert the "correction" string into
SQL that is executed to perform the calculation.
Name
convert_conc — convert a quantity from one given unit to another.
Synopsis
NUMERIC
convert_conc
( | quantity, | |
| from_unit, | ||
to_unit); |
NUMERIC
quantity
;
TEXT
from_unit
;
TEXT
to_unit
;Input
quantity
The concentration in need of conversion, e.g. a NUCACID_CONC_DATA.Quantity value. A NUMERIC value, or any other data type that can be interpreted as a number and converted to NUMERIC.
from_unit
A text string indicating the unit to convert from. In other words, the current unit of the quantity. This must be a NUCACID_CONC_UNITS.Unit value.
to_unit
A text string indicating the unit to convert into. In other words, the desired unit of the quantity. This must be a NUCACID_CONC_UNITS.Unit value.
Description
Returns a number, the converted concentration. If
the two units are not related — their respective
NUCACID_CONC_UNITS.Reference values are not
equal — then no conversion can occur and this
function returns NULL.
This function is used by the NUCACIDS_W_CONC view to generate its several concentration columns. It may also be useful to users trying to standardize concentrations when querying NUCACID_CONC_DATA.
Name
to_hhmmss — convert an interval into a string with format
"HH:MM:SS". E.g. 1 day 02:03:04
becomes 26:03:04.
Synopsis
TEXT
to_hhmmss
( | this_interval); |
INTERVAL
this_interval
;Description
Returns a text string, the interval converted into HH:MM:SS format.
This function is used by the CURATED_TEMPS view to standardize the format of the Timespan and Time_Missed columns.
Caution
Intervals that include time units of variable length like months and years will yield unpredictable results.
The logout program logs the user out of the Babase web based collection of programs.
Note
Babase consists of many programs, only some of which are web based programs written specifically for Babase. The logout program only controls access to those programs written specifically for Babase, other “off the shelf” programs have their own logout mechanisms.
Note
Logout from Babase is automatic after a period of inactivity.[305]
The wwwdiff program compares two text files. It can be found on the Babase Web site.
Among other uses, this program provides a useful data validation mechanism. To validate data, have two different individuals enter the data and compare the results with the wwwdiff program. It is unlikely that both individuals will make identical errors and so almost all data transcription errors should be caught using this method.
Tip
The program uploads the two files to be compared. For security reasons most web browsers will always clear the names of the uploaded files once they have been uploaded. This makes it difficult to repeatedly upload the same or similar files, as when re-comparing two files after correcting errors. The situation is not as bad as it might sound because browsers will often provide a “browse” button and keep track of the directory last accessed, removing the need to re-navigate to the location of the data files. But it is still awkward to have to repeatedly point and click.
One solution is to use the browser's “reload” button. This will repeat the upload and comparison of the two files, but using the new, corrected, file content. A second, less desirable possibility is to have the have the pathnames of the files handy in a text document and cut and paste them as needed. A third possibility might be to use the browser's “back” button, but browsers will often clear the file upload information in this case in the same fashion they would with password information.
The wwwdiff program provides 5 comparison methods:
- Tabular by Word -- Suppress identical lines
Like Tabular by Word, below, but identical lines are not displayed.
- Tabular by Word
Useful where the data consists of individual words aligned as rows and columns of data. Compares the files contents on a word-by-word basis and displays the entire content of both files as a table with differences marked.
Caution
When one file contains whitespace[306] that is not in the other, this comparison method shows extra cells in the output. Words are separated by whitespace but because there is only extra whitespace the cells are empty. Thus, when one file contains more whitespace than the other those rows will contain more columns. Normally the data in the extra cells would be color coded to inform as to whence they came, but because the cells are empty there is nothing to color code. The operator must compare the files by hand to determine which file contains the extra whitespace.
- By Word
Useful where the data consists of words and there are relatively few changes or where paragraphs have been refilled and words have moved from line to line. Compares the file contents on a word-by-word basis displaying the entire content of both files as plain text.
- By Line
Useful when there is a large amount of textual data. Compares the file contents on a line-by-line basis and displays only a small amount of context surrounding those portions of the text which differ between the files.
- By File
Useful when comparing non-text files. Reports the location (by line and byte offset from the beginning of the files) of the first difference found.
Note
When comparing using any of the “by word” methods the reported line number increases by one when either: File A or File B contains an entirely new line not in the other file, or when a line in one file differs in every word from the same line in the other file. This throws the line numbering off relative to one or both of the original files.
These procedures provide a mechanism for manually updating the analyzed data which Babase maintains. They are expected to be of use only to the data managers.
As a rule the analyzed data are kept up-to-date automatically by Babase (the exceptions are the CYCSTATS and the REPSTATS tables, and the MEMBERS.Supergroup and residency columns on MEMBERS), but at times it may be necessary to reconstruct the analyzed data. One such occasion would be the discovery of a bug in the Babase code which keeps the analyzed data up-to-date.
The procedures tend to come in pairs, one of which updates an entire table and the other of which updates only the data related to a specific Sname.
Table 8.2. Data Analysis Procedures
| Name | Description |
|---|---|
|
rebuild the automatic Mdates for an individual | |
|
rebuild the automatic Mdates of all individuals | |
|
rebuild the table for an individual | |
|
rebuild the entire table | |
|
rebuild the table for an individual | |
|
rebuild the entire table | |
|
rebuild the table for an individual | |
|
rebuild the entire table | |
|
re-interpolate the MEMBERS table for an individual, re-construct the Supergroup and Delayed_Supergroup columns, and re-analyze the residency information for the individual's MEMBERS rows | |
|
re-interpolate the MEMBERS table for all individuals, re-construct the Supergroup and Delayed_Supergroup columns, and re-analyze the residency information for all individuals' MEMBERS rows | |
|
for all individuals with any rows that haven't had their supergroups constructed or residency analyzed, re-interpolate all their MEMBERS rows, re-construct all their Supergroup and Delayed_Supergroup columns, and re-analyze the residency information for all their MEMBERS rows | |
|
rebuild the table for an individual | |
|
rebuild the entire table | |
|
rebuild the table for an individual | |
|
rebuild the entire table | |
|
rebuild the calculated columns in RANKS for all rows with a specific Grp, Rnkdate, and Rnktype | |
|
rebuild the calculated columns in RANKS for all rows with a specific Grp and Rnktype | |
|
rebuild the calculated columns in the entire table | |
|
rebuild the table for an individual | |
|
rebuild the entire table | |
|
rebuild the residency related columns of the MEMBERS table and repopulate the RESIDENCIES table for an individual | |
|
rebuild the residency related columns of the MEMBERS table and repopulate the entire RESIDENCIES table for all individuals | |
|
rebuild the residency-related columns of the MEMBERS table and repopulate the entire RESIDENCIES table, but only for the individuals with MEMBERS rows that haven't already been analyzed | |
|
rebuild the table for an individual | |
|
rebuild the entire table | |
|
rebuild the Supergroup and Delayed_Supergroup columns of the MEMBERS table for an individual | |
|
rebuild the Supergroup and Delayed_Supergroup columns of the entire MEMBERS table | |
|
rebuild the Supergroup and Delayed_Supergroup columns of the entire MEMBERS table for all individuals who have any rows that have not had their Supergroup or Delayed_Supergroup built | |
|
Rapidly delete old style CENSUS rows |
Name
rebuild_automdates — rebuild the automatic Mdates for an individual
Synopsis
int
rebuild_automdates
( | sname); |
char(3)
sname
;Description
This procedure rebuilds the automatic Mdates on the CENSUS table for a specific individual.
Warning
This routine should not be run while triggers (automatic data validation) are enabled.
Example
-- Turn off triggers by e.g.:
-- cd db/triggers
-- make BABASE_DB=babase_foo destroy
BEGIN;
-- Empty those tables that could refer to automdates.
DELETE FROM cycstats WHERE sname = 'FOO';
DELETE FROM mmintervals WHERE sname = 'FOO';
DELETE FROM mdintervals WHERE sname = 'FOO';
SELECT rebuild_all_automdates();
-- Commit so that triggers can be re-installed.
COMMIT;
-- Re-install trigges by e.g.:
-- cd db/triggers
-- make BABASE_DB=babase_foo install
BEGIN;
-- Rebuild the cycles.seq and cycles.series columns
SELECT rebuild_cycles('FOO');
-- Rebuild the tables temporarly emptied.
SELECT rebuild_cycstats('FOO');
SELECT rebuild_mmintervals('FOO');
SELECT rebuild_mdintervals('FOO');
COMMIT;
Name
rebuild_all_automdates — rebuild the automatic Mdates of all individuals
Synopsis
int
rebuild_all_automdates
( | ); |
Description
This procedure rebuilds the automatic Mdates of all individuals on the CENSUS table.
Warning
This routine should not be run while triggers (automatic data validation) are enabled.
Example
-- Turn off triggers by e.g.:
-- cd db/triggers
-- make BABASE_DB=babase_foo destroy
BEGIN;
-- Empty those tables that could refer to automdates.
DELETE FROM cycstats;
DELETE FROM mmintervals;
DELETE FROM mdintervals;
SELECT rebuild_all_automdates();
-- Commit so that triggers can be re-installed.
COMMIT;
-- Re-install trigges by e.g.:
-- cd db/triggers
-- make BABASE_DB=babase_foo install
BEGIN;
-- Rebuild the cycles.seq and cycles.series columns
SELECT rebuild_all_cycles();
-- Rebuild the tables temporarly emptied.
SELECT rebuild_all_cycstats();
SELECT rebuild_all_mmintervals();
SELECT rebuild_all_mdintervals();
COMMIT;
Name
rebuild_cycgapdays — rebuild the CYCGAPDAYS table for an individual
Synopsis
int
rebuild_cycgapdays
( | sname); |
char(3)
sname
;Description
This procedure rebuilds the CYCGAPDAYS table, using the CYCGAPS table as its source, for a specific individual.
Name
rebuild_all_cycgapdays — rebuild the entire CYCGAPDAYS table
Synopsis
int
rebuild_all_cycgapdays
( | ); |
Description
This procedure rebuilds the entire CYCGAPDAYS table, using the CYCGAPS table as its source.
Name
rebuild_cycles — rebuild the CYCLES table for an individual
Synopsis
int
rebuild_cycles
( | sname); |
char(3)
sname
;Description
This procedure rebuilds the CYCLES table for a specific individual. The Mdates, Tdates, and Ddates for the individual are collected into cycles and the Seq and Series are re-computed. The CYCPOINTS and CYCGAPS tables, as well as the CYCLES table itself, provide the data necessary to rebuild CYCLES.
Name
rebuild_all_cycles — rebuild the entire CYCLES table
Synopsis
int
rebuild_all_cycles
( | ); |
Name
rebuild_cycstats — rebuild the CYCSTATS table for an individual
Synopsis
int
rebuild_cycstats
( | sname); |
char(3)
sname
;Name
rebuild_all_cycstats — rebuild the entire CYCSTATS table
Synopsis
int
rebuild_all_cycstats
( | ); |
Name
rebuild_members — re-interpolate the MEMBERS table for an individual, re-construct the Supergroup and Delayed_Supergroup columns, and re-analyze the residency information for the individual's MEMBERS rows
Synopsis
int
rebuild_members
( | sname); |
char(3)
sname
;Description
This procedure re-interpolates the MEMBERS table for a specific individual. The CENSUS table provides the data necessary to rebuild MEMBERS.
The MEMBERS.Supergroup and Delayed_Supergroup columns and the residency columns on MEMBERS are also re-computed.[307]
[307] See: rebuild_supergroup() and rebuild_residency()
[308] Copying an individual's CENSUS rows into MEMBERS can be accomplished with code like the following:
BEGIN;
-- First remove existing census-like rows for individual "FOO"
DELETE FROM members
WHERE members.sname = 'FOO'
AND members.origin <> 'I';
-- Then copy rows from CENSUS to MEMBERS.
INSERT INTO members (sname, date, grp, origin, interp)
SELECT census.sname, census.date, census.grp, census.status, 0
FROM census
WHERE census.sname = 'FOO'
AND census.status <> 'A';
COMMIT;
Name
rebuild_all_members — re-interpolate the MEMBERS table for all individuals, re-construct the Supergroup and Delayed_Supergroup columns, and re-analyze the residency information for all individuals' MEMBERS rows
Synopsis
int
rebuild_all_members
( | ); |
Description
This procedure re-interpolates the entire MEMBERS table. The CENSUS table provides the data necessary to rebuild MEMBERS.
The MEMBERS.Supergroup and Delayed_Supergroup columns and the residency columns for all MEMBERS rows are also re-computed.[309]
Caution
“rebuild_members” is something of a misnomer because the program assumes that MEMBERS already contains all the (non-absent) CENSUS rows. See rebuild_members().
Name
rebuild_new_members — for all individuals with any rows that haven't had their supergroups constructed or residency analyzed, re-interpolate all their MEMBERS rows, re-construct all their Supergroup and Delayed_Supergroup columns, and re-analyze the residency information for all their MEMBERS rows
Synopsis
int
rebuild_new_members
( | ); |
Description
This procedure queries MEMBERS
to determine which individuals have any rows with NULL
Supergroup, Delayed_Supergroup, or Residency, and then re-interpolates
all of the MEMBERS rows for those
individuals. The CENSUS table provides
the data necessary to rebuild the MEMBERS rows.
The MEMBERS.Supergroup and Delayed_Supergroup columns and the residency columns for all those individuals' MEMBERS rows are also re-computed.[310]
Caution
“rebuild_members” is something of a misnomer because the program assumes that MEMBERS already contains all the (non-absent) CENSUS rows. See rebuild_members().
Name
rebuild_mdintervals — rebuild the MDINTERVALS table for an individual
Synopsis
int
rebuild_mdintervals
( | sname); |
char(3)
sname
;Description
This procedure rebuilds the MDINTERVALS table for a specific individual. The CYCLES, CYCPOINTS, and CYCGAPS tables provide the data necessary to rebuild MDINTERVALS.
Note
The MDINTERVALS table is not automatically maintained by the system at this time.
Name
rebuild_all_mdintervals — rebuild the entire MDINTERVALS table
Synopsis
int
rebuild_all_mdintervals
( | ); |
Description
This procedure rebuilds the entire MDINTERVALS table. The CYCLES, CYCPOINTS, and CYCGAPS tables provide the data necessary to rebuild MDINTERVALS.
Note
The MDINTERVALS table is not automatically maintained by the system at this time.
Name
rebuild_mmintervals — rebuild the MMINTERVALS table for an individual
Synopsis
int
rebuild_mmintervals
( | sname); |
char(3)
sname
;Description
This procedure rebuilds the MMINTERVALS table for a specific individual. The CYCLES, CYCPOINTS, and CYCGAPS tables provide the data necessary to rebuild MMINTERVALS.
Note
The MMINTERVALS table is not automatically maintained by the system at this time.
Name
rebuild_all_mmintervals — rebuild the entire MMINTERVALS table
Synopsis
int
rebuild_all_mmintervals
( | ); |
Description
This procedure rebuilds the entire MMINTERVALS table. The CYCLES, CYCPOINTS, and CYCGAPS tables provide the data necessary to rebuild MMINTERVALS.
Note
The MMINTERVALS table is not automatically maintained by the system at this time.
Name
rebuild_ranks — rebuild the calculated columns in RANKS for all rows with a specific Grp, Rnkdate, and Rnktype
Synopsis
int
rebuild_ranks
( | grp, | |
| rnkdate, | ||
rnktype); |
NUMERIC(6,4)
grp
;
date
rnkdate
;
varchar(6)
rnktype
;Description
This procedure rebuilds the Ags_Density, Ags_Reversals, and Ags_Expected columns for all rows in RANKS with the specified Grp, Rnkdate, and Rnktype. The INTERACT_DATA, PARTS, and RANKS tables provide the data necessary to rebuild these columns.
Note
These columns are not automatically maintained by the system at this time.
Name
rebuild_ranks_grp_rnktype — rebuild the calculated columns in RANKS for all rows with a specific Grp and Rnktype
Synopsis
int
rebuild_ranks_grp_rnktype
( | grp, | |
rnktype); |
NUMERIC(6,4)
grp
;
varchar(6)
rnktype
;Description
This procedure rebuilds the Ags_Density, Ags_Reversals, and Ags_Expected columns for all rows in RANKS with the specified Grp and Rnktype. The INTERACT_DATA, PARTS, and RANKS tables provide the data necessary to rebuild these columns.
Note
These columns are not automatically maintained by the system at this time.
Name
rebuild_all_ranks — rebuild the calculated columns in the entire RANKS table
Synopsis
int
rebuild_all_ranks
( | ); |
Description
This procedure rebuilds the Ags_Density, Ags_Reversals, and Ags_Expected columns in the entire RANKS table. The INTERACT_DATA, PARTS, and RANKS tables provide the data necessary to rebuild these columns.
Note
These columns are not automatically maintained by the system at this time.
Name
rebuild_repstats — rebuild the REPSTATS table for an individual
Synopsis
int
rebuild_repstats
( | sname); |
char(3)
sname
;Name
rebuild_all_repstats — rebuild the entire REPSTATS table
Synopsis
int
rebuild_all_repstats
( | ); |
Name
rebuild_residency — rebuild the residency related columns of the MEMBERS table and repopulate the RESIDENCIES table for an individual
Synopsis
int
rebuild_residency
( | sname); |
char(3)
sname
;Description
This procedure rebuilds the residency related columns of the MEMBERS table for a specific individual. These are:
Also, the individual's rows in the RESIDENCIES table (if any) are emptied and replaced.
The interpolated and Supergroup information stored in MEMBERS, and BIOGRAPH, provide the data necessary to rebuild residency.
Caution
The MEMBERS.Supergroup and Delayed_Supergroup columns must be updated[312] if residency is to be correctly computed.
Note
The residency information is not automatically maintained by the system at this time.
[311] Technically, MEMBERS.LowFrequency is an independent computation from the residency information. However it is convenient to compute it along with the residency columns.
Name
rebuild_all_residency — rebuild the residency related columns of the MEMBERS table and repopulate the entire RESIDENCIES table for all individuals
Synopsis
int
rebuild_all_residency
( | ); |
Description
This procedure rebuilds the residency related columns of the MEMBERS table for all individuals. All data in the RESIDENCIES table is also updated appropriately. The interpolated and Supergroup information stored in MEMBERS, and BIOGRAPH, provide the data necessary to rebuild residency.
Caution
The MEMBERS.Supergroup and Delayed_Supergroup columns must be updated[313] if residency is to be correctly computed.
Note
The residency information is not automatically maintained by the system at this time.
Name
rebuild_new_residency — rebuild the residency-related columns of the MEMBERS table and repopulate the entire RESIDENCIES table, but only for the individuals with MEMBERS rows that haven't already been analyzed
Synopsis
int
rebuild_new_residency
( | ); |
Description
This procedure queries MEMBERS
to determine which individuals have any rows with NULL
Residency, and then
re-analyzes the residency related
columns for all of the MEMBERS rows for those individuals. Those
individuals' data in the RESIDENCIES
table will also be updated appropriately. The interpolated
and Supergroup information
stored in MEMBERS, and BIOGRAPH, provide the data necessary to
rebuild residency.
Caution
The MEMBERS.Supergroup and Delayed_Supergroup columns must be updated[314] if residency is to be correctly computed.
Note
The residency information is not automatically maintained by the system at this time.
Name
rebuild_sexskins — rebuild the SEXSKINS table for an individual
Synopsis
int
rebuild_sexskins
( | sname); |
char(3)
sname
;Description
This procedure rebuilds the SEXSKINS table for a specific individual in that it re-associates an individuals SEXSKINS rows with the correct sexual cycle as explained in the Sexual Cycle Determination section. The CYCLES, and CYCPOINTS tables provide the data necessary to rebuild SEXSKINS.
Name
rebuild_all_sexskins — rebuild the entire SEXSKINS table
Synopsis
int
rebuild_all_sexskins
( | ); |
Description
This procedure rebuilds the SEXSKINS table for a specific individual in that it re-associates an individuals SEXSKINS rows with the correct sexual cycle as explained in the Sexual Cycle Determination section. The CYCLES, and CYCPOINTS tables provide the data necessary to rebuild SEXSKINS.
Name
rebuild_supergroup — rebuild the Supergroup and Delayed_Supergroup columns of the MEMBERS table for an individual
Synopsis
int
rebuild_supergroup
( | sname); |
char(3)
sname
;Description
This procedure rebuilds the Supergroup and Delayed_Supergroup columns of the MEMBERS table for a specific individual. The GROUPS, CENSUS (by way of MEMBERS), and BIOGRAPH tables provide the necessary data.
If this individual has any MEMBERS rows with non-NULL Residency, LowFrequency, or GrpOfResidency, those values
depended on an earlier version of the supergroup data,
so this function automatically sets them all to NULL
and removes any of the individual's rows in the RESIDENCIES table.
Note
The MEMBERS.Supergroup and Delayed_Supergroup columns are not automatically maintained by the system at this time.
Name
rebuild_all_supergroup — rebuild the Supergroup and Delayed_Supergroup columns of the entire MEMBERS table
Synopsis
int
rebuild_all_supergroup
( | ); |
Description
This procedure rebuilds the Supergroup and Delayed_Supergroup columns on the entire MEMBERS table. The GROUPS, CENSUS (by way of MEMBERS), and BIOGRAPH tables provide the necessary data.
If there are any MEMBERS rows
with non-NULL Residency,
LowFrequency, or GrpOfResidency, those values
depended on an earlier version of the supergroup data,
so this function automatically sets them all to NULL
and removes all rows in the RESIDENCIES table.
Note
The Supergroup and Delayed_Supergroup columns of the MEMBERS table are not automatically maintained by the system at this time.
Name
rebuild_new_supergroup — rebuild the Supergroup and Delayed_Supergroup columns of the entire MEMBERS table for all individuals who have any rows that have not had their Supergroup or Delayed_Supergroup built
Synopsis
int
rebuild_new_supergroup
( | ); |
Description
This procedure queries MEMBERS
to determine which individuals have any rows with NULL
Supergroup or Delayed_Supergroup, and then
rebuilds the Supergroup and
Delayed_Supergroup columns for
all of the MEMBERS rows for those individuals. The
GROUPS, CENSUS (by
way of MEMBERS), and BIOGRAPH tables provide the necessary
data.
If the individuals have any MEMBERS rows with non-NULL Residency, LowFrequency, or GrpOfResidency, those values
depended on an earlier version of the supergroup data,
so this function automatically sets them all to NULL
and removes any of those individuals' rows in the RESIDENCIES table.
Note
The Supergroup and Delayed_Supergroup columns of the MEMBERS table are not automatically maintained by the system at this time.
Name
delete_census — Rapidly delete “old style” CENSUS rows
Synopsis
int
delete_census
( | sname, | |
| from, | ||
through); |
char(3)
sname
;
date
from
;
date
through
;Description
A function to delete an individual's rows from CENSUS for a particular time period. The deleted rows are inclusive of the supplied dates.
This function is useful because deleting multiple non-interpolating CENSUS rows in a single SQL DELETE statement is an operation that takes an amount of time proportional to the square of the number of rows deleted. Use of this function is a substitute for deleting CENSUS rows one at a time.
Note
Eventually most of the non-interpolating CENSUS rows will be removed from Babase, along with their codes. These are the rows associated with analysis of historical data.
Tip
To delete more than a years worth of census data it's best to delete a year at a time, and leave a single row undeleted within each year. When done go back and delete the single rows. This can all be done by submitting multiple statements at once so as not to have to continually interact with the system. Not only will this technique minimize the time spent it will also minimize the number of MEMBERS rows created and destroyed, and therefore the number of MEMBERS.Membids used.
Example
-- Deleting CENSUS rows for FOO from 1987-03-18
-- through 1992-01-23, inclusive.
BEGIN TRANSACTION;
SELECT delete_census('FOO', '1987-03-18', '1988-03-17');
SELECT delete_census('FOO', '1988-03-19', '1989-03-17');
SELECT delete_census('FOO', '1989-03-19', '1990-03-17');
SELECT delete_census('FOO', '1990-03-19', '1991-03-17');
SELECT delete_census('FOO', '1991-03-19', '1992-01-23');
DELETE FROM census
WHERE census.sname = 'FOO' AND census.date = '1988-03-18';
DELETE FROM census
WHERE census.sname = 'FOO' AND census.date = '1989-03-18';
DELETE FROM census
WHERE census.sname = 'FOO' AND census.date = '1990-03-18';
DELETE FROM census
WHERE census.sname = 'FOO' AND census.date = '1991-03-18';
COMMIT TRANSACTION;
[294] Uploading into a single row of a view can update multiple tables, and programs designed to handle specialized input data formats may update arbitrary portions of the database as needed.
[295] Once an error occurs no changes will be committed to the database.
[296] This check will not detect duplicate darting logistic data because uploading darting logistic data creates new rows in DARTINGS.
[297] Note that the test is against the text of the
sname and dartdate
as entered in the uploaded file, not, e.g., the actual
date. So this test fails when the same date is written in
two different, but valid, forms.
[298] Alternately, as usual, the uploaded cells can be empty and nothing will be added to the database.
[300] See PID_String
[301] See SID_String
[302] This may or may not result in a NULL value in
the database, depending on how the program is
invoked.
[303] Those that end in “_func” are the procedures used by the triggers for data validation.
[304] Yes, %s is ugly, but this was
not chosen arbitrarily. It has a very specific meaning
for the PostgreSQL FORMAT() function, which is used in
this function to convert the "correction" string into
SQL that is executed to perform the calculation.
[305] Currently 1 hour.
[306] Spaces, tabs, and whatever other characters that can't been seen.
[307] See: rebuild_supergroup() and rebuild_residency()
[308] Copying an individual's CENSUS rows into MEMBERS can be accomplished with code like the following:
BEGIN;
-- First remove existing census-like rows for individual "FOO"
DELETE FROM members
WHERE members.sname = 'FOO'
AND members.origin <> 'I';
-- Then copy rows from CENSUS to MEMBERS.
INSERT INTO members (sname, date, grp, origin, interp)
SELECT census.sname, census.date, census.grp, census.status, 0
FROM census
WHERE census.sname = 'FOO'
AND census.status <> 'A';
COMMIT;
[309] See: rebuild_supergroup() and rebuild_residency()
[310] See: rebuild_supergroup() and rebuild_residency()
[311] Technically, MEMBERS.LowFrequency is an independent computation from the residency information. However it is convenient to compute it along with the residency columns.
Because time values in focal sampling data record the time the observation was entered, which necessarily occurs after the observation is taken, it may be desirable to ignore the additional seconds. The views and functions that Babase supplies to do this are documented elsewhere. This appendix provides an introduction to the underlying PostgreSQL facilities supporting these sorts of operations.
The PostgreSQL date_trunc()
function can be used to produce a time with the seconds forced to
0. Here is an example:
Example A.1. Using the Postgresql date_trunc() function to set
seconds to zero
babase=> select date_trunc('minute', '23:15:52'::time);
date_trunc
------------
23:15:00
(1 row)
To obtain the portion of timestamp that the
date_trunc() function discards, use the Babase
date_mod() function, which defines
date_trunc(period, daytime) as daytime -
date_trunc(period, daytime).
Example A.2. Using the Babase date_mod() function to
return the minutes and seconds.
babase=> select date_mod('hour', '23:15:52'::time);
date_mod
------------
00:15:52
(1 row)
The date_trunc() function produces a time,
which is a suitable sort of value for further computation, the
calculation of intervals, etc. To produce human readable text in
the form HH:MM the PostgreSQL to_char()
function may be used.
Example A.3. Using the Postgresql to_char() function to
convert times to HH:MM text
babase=> select to_char('23:15:52'::time, 'HH24:MI');
to_char
---------
23:15
(1 row)
For further information computations which may be performed using dates and times see the PostgreSQL documentation on Date/Time Functions and Operators.
There are many ways to query for all-occurrences interactions. This appendix utilizes the PostgreSQL EXISTS() function, which is useful when the result of the query does not need any columns from the SAMPLES table. A regular join would work as well.
Example B.1. Finding all the all-occurrences interactions
SELECT *
FROM actor_actees
WHERE EXISTS(SELECT 1
FROM samples
WHERE samples.sid = actor_actees.sid
AND (samples.sname = actor_actees.actor
OR samples.sname = actor_actees.actee);
There are some circumstances in which the id of a sexual cycle (Cid) must be altered. This appendix presents one example showing what happens when sexual cycle events are added to the database. Similar alterations are required when sexual cycle events are deleted, a record of a gap in observation is added to the database, or a record of a gap in observation is removed from the database.
Note
The tables shown in the example contain some, but not all, of the columns of both CYCLES and CYCPOINTS.
Example C.1. “Splitting” a sexual cycle in two
Suppose there is, in date order, a Mdate, Tdate, and
Ddate. They are all in the same cycle, and so have the same
Cid. (Say Cid 10. Consequently they have the
same CYCLES.Seq, say Seq 1).
Now, a new Tdate, Ddate, and Mdate (in that order by date) are
added to the database. Their dates all fall between the Mdate
and Tdate with Cid 10. The result is:
Table C.2. Sexual cycle events after insertion
| Cid | Seq | Code | Date |
|---|---|---|---|
11 | 1 | M | Date 1 |
11 | 1 | T | Date 1.1 |
11 | 1 | D | Date 1.2 |
10 | 2 | M | Date 1.3 |
10 | 2 | T | Date 2 |
10 | 2 | D | Date 3 |
The first Mdate, Date 1, has changed its Cid. Dates 2 and 3,
the original Tdate and Ddate, have changed their Seq.
Although this sort of thing may only happen when mistakes are corrected, when it does happen there's no way around changing some Mdate, Tdate, or Ddate's Cid.
Table of Contents
Babase 5.0 was released on May 3, 2022. In this update, all tables (except for those related to warnings) became "temporal" tables. All changes to the data from this date onward will be recorded, in case earlier versions need to be recalled.
Major changes to Babase 5.0 include:
Creation of the babase_history schema and its "history" tables
Addition of a Sys_Period column to every table
Installation of "versioning" triggers to record old versions of data in the "history" tables
For information about how to use this new system, see Temporal Tables and babase_history.
Babase 5.1 was released on June 2, 2022. In this update, three new columns were added to RANKS to assist users with making decisions about the accuracy of rank data, similar to the way "confidence" columns are employed in other tables. New functions were also added to automate the population of these new columns.
Major changes in Babase 5.1 include:
Addition of the Ags_Density, Ags_Reversals, and Ags_Expected columns to the RANKS table
Creation of the rebuild_ranks(), rebuild_ranks_grp_rnktype(), and rebuild_all_ranks() functions to populate the new columns
In Babase 5.1.1 — released on June 16, 2022 — most of the validation for the CYCGAPS table was rewritten. For the most part, the actual rules did not change (exceptions discussed below) but the timing did, such that most of the CYCGAPS rules are now validated on transaction commit. Because of this change, rules that were in place to allow adding/removing/modifying gaps by doing odd things — e.g. "insert a 'start' row with the same date as an 'end' row to remove both" — are no longer needed and were removed.
Babase 5.2 was released on August 9, 2022. In this update, the contents of the PCSKINS table were added to the SEXSKINS table, and PCSKINS was removed. The PCSKINS_SORTED view was likewise removed. To facilitate this change, the Color column was added to SEXSKINS.
In Babase 5.2.1 — released on August 15, 2022 — the REPRO_NOTES table was added, as well as the SEXSKINS_REPRO_NOTES view.
In Babase 5.2.2 — released on September 7, 2022 — the Comments column was added to the WBC_COUNTS table.
In Babase 5.2.3 — released on September 8, 2022
— the HYBRIDGENE_SCORES table was
altered to allow Lower_Conf and Upper_Conf to be NULL.
Babase 5.3 was released on June 21, 2023. In this update, the residency system was revamped. Details of the new system are explained in the residency rules, but in general:
Individuals no longer need to be present in a group for 29 days before their residency begins. Instead, their residency begins on the first day that they were continually present in the group of residency.
When an individual begins to transition out of their resident group, their residency no longer ends at the beginning of that transition period (their first absence). Instead, residency lasts until the end of that transition period (the last day of the last "present" 29-day window).
Residency assignments are no longer limited to study groups. Individuals can be residents of any group (except 9.0 and 10.0), and it is possible to remain resident in a low-frequency (probably non-study) group through lengthy periods of nonobservation.
The system no longer attempts to assign residency in all of an individual's MEMBERS rows. Instead, the system only considers dates between the individual's Entrydate and Statdate, inclusive.
Other changes in Babase 5.3:
A new column added to ENTRYTYPES, telling the system when to use an alternate rule set for assigning residency on and shortly after an individual's Entrydate.
A new column added to STATUSES, telling the system when to use an alternate rule set for assigning residency on and shortly before an individual's Statdate.
A new table, RESIDENCIES, which condenses each individual's day-by-day residency information in MEMBERS into discrete "bouts".
In Babase 5.3.1, released on 17 July 2023, the FLOW_CYTOMETRY table was added.
In Babase 5.3.2, released on 02 November 2023, the Collection_Date_Status column was added to TISSUE_DATA and several related views.
In Babase 5.3.3, released on 09 November 2023, the VAGINAL_PHS table and the related VAGINAL_PH_STATS view were added.
Babase 5.4 was released on November 28, 2023. In this update, the WEATHERHAWK table was renamed to DIGITAL_WEATHER, the Lightning_Strikes column was added to DIGITAL_WEATHER, and the WEATHERHAWK_SOFTWARES table was renamed to WEATHER_SOFTWARES.
Babase 5.5 was released on January 26, 2024. In this update, most of the validation of focal sampling data was rewritten.
Previously, two specific sample types (F for adult females and J for juveniles of any sex) were hard coded into the system as the only legal SAMPLES.SType values. When a rule applied to one of those STypes but not the other, the rule was also hard coded into the system. All of that hard coding was removed in this update.
Instead, the STYPES, STYPES_ACTIVITIES, STYPES_NCODES, and STYPES_POSTURES tables were added. They have been populated so that all previously hard coded rules are still being enforced. In other words, the data and validation on it did not change (with one exception, below). Rather, the way that the validation is written and enforced has changed. This change should allow the system to add focal sampling data that were collected with other sampling protocols.
As mentioned above, there is one exception to the claim that the "data and validation on it did not change". Before this version, a female could not be sampled as a juvenile after the conception date of her first offspring. This rule was deemed unnecessarily restrictive. Now, a female can be sampled as a juvenile until the birth date of her first offspring, thanks to the STYPES.Days_After_FirstBirth column.
In Babase 5.5.1 — released on
February 09, 2024 — the DIGITAL_WEATHER table was adjusted to accommodate
rainfall measurements from devices besides the WeatherHawk.
The YearlyRain column is now
allowed to be NULL, and the automatic calculation of TimeStampRain from the YearlyRain was removed.
In Babase 5.5.2, released on 22 February 2024, the PALMTOPS support table was renamed to SAMPLES_COLLECTION_SYSTEMS, and the SAMPLES.Palmtop column was renamed to Collection_System. The rationale for this change is discussed in the documentation of the SAMPLES_COLLECTION_SYSTEMS table.
In Babase 5.5.3, released on 06 Mar 2024, a few more adjustments were made to the DIGITAL_WEATHER table. Barometer was changed to contain corrected values, not uncorrected ones. This included a slight change in the range of allowed values and the expansion of the column's data type to allow another digit. Also, the TimeStampRain was converted from an integer to a 5-digit number with 2 digits to the right of the decimal.
In Babase 5.5.4, released on 22 Apr 2024, the SWERB views learned about longitude and latitude. Specifically, the SWERB, SWERB_DATA_XY, SWERB_DEPARTS, SWERB_GW_LOC_DATA_XY, SWERB_GW_LOCS, and SWERB_LOC_GPS_XY views added longitude and latitude colums alongside the x and y columns they already had, and the SWERB_UPLOAD view became able to upload data with longitude and latitude coordinates. The bb_makepoint_longlat() function was added, as well.
In Babase 5.5.5, released on 31 May
2024, the INTERACT_DATA table was changed so
that it no longer requires that interactions with a non-NULL
Sid must have a Handwritten of FALSE. Whether or
not an interaction is associated with a focal sample is now
fully independent of whether or not it was recorded by hand.
Admittedly, it could be argued that this is one of the "hard
coded" rules that should have been removed in Babase 5.5. In this case the rule is not being
replaced or enforced elsewhere; it is simply being
removed.
Babase 5.6 was released on January 14, 2025. In this update, the tables in the inventory were changed so that there are more options for recording a sample's source(s). Specifically:
The TISSUE_SOURCES table was added, allowing us to record when tissue samples come from other tissues.
The NUCACID_SOURCES table now allows a nucleic acid sample to have more than one row, i.e. more than one source.
The TISSUE_DATA.Multi_Indivs column was added, including new rules for situations where a sample might not have a UIId.
Nucleic acid samples no longer solely depend on their source TId to identify their source individual. The UIId column was added to NUCACID_DATA.
The NUCACID_DATA.Multi_Indivs and Multi_TIds columns were added, including new rules for situations where a sample might not have a UIId and/or a TId.
The NUCACID_SOURCES_EXT and TISSUE_SOURCES_EXT views were added, making it easier to see the LocalIds of the samples and their sources.
In Babase 5.6.1, released on 30 Jan 2025, several views were added to facilitate the upload of data from dartings. Specifically, these views:
In Babase 5.6.2, released on 05 Feb 2025, the MPIACTS table's "Multi_First" column was removed, along with the validation that it enforced. This column was poorly enforcing a rule that we didn't actually want enforced at all, so it needed to go.
In Babase 5.6.3, released on 12 Mar 2025, the ability to identify interactions as "representative" was added via the new ACTOR_ACTEES column: Repr_Interxn. Related to that, the STYPES table also added a new column, Repr_Interxns.
Babase 5.7 was released on March 24, 2025. In this update, the old system for recording paternities was discarded. It was effective at recording paternities that were determined from microsatellite genotyping and analyzed by a specific analysis software, but it was essentially useless for recording paternities that were determined any other way. The system was replaced with a new set of tables that are more robust to methodological changes in paternity analyses. Specifically:
The DADS_ANALYSES, DADS_CONSENSUS, DADS_EVIDENCE, and DADS_EVIDENCE_TERMS tables were added.
Tables formerly known as DAD_DATA_COMPLETENESS and DAD_DATA_MISMATCHES were renamed to DADS_COMPLETENESS and DADS_MISMATCHES, respectively.
The PARENTS view was adjusted. It now gets its Dad column from DADS_CONSENSUS.Dad_Consensus and no longer has a Dadid column.
The old DAD_DATA and DAD_SOFTWARE tables were dropped. Their data were added to the new tables, so no data were lost.
In Babase 5.7.1, released on 16 May 2025, the hybrid morphology scores were added. Specifically, the HYBRIDMORPH_OBSERVERS, HYBRIDMORPH_REPORTS, and HYBRIDMORPH_SCORE_DATA tables and the HYBRIDMORPH_SCORES view were created and populated.
In Babase 5.7.2, released on 02 Jul 2025, the multiparty interactions gained the ability to record their observer. The MPIS table added an Observer column, which also now appears in MPI_EVENTS and which can be populated by the MPI_UPLOAD view.
In Babase 5.7.3, released on 12 Aug 2025, the Supergroup column was revised (or rather, the function that populates it was revised) for a particular rare circumstance. When the Grp has no Permanent date, the Supergroup should be that group's From_group. When this occurs now, the system will check if that From_Group has ceased to exist. If it no longer exists, then the Supergroup cannot be the From_Group and is instead set to the Grp.
In Babase 5.7.4, released on 28 Aug 2025, several new columns were added to DIGITAL_WEATHER to allow it record temperatures from a second temperature probe. Specifically, the Secondary_Temp_Probe, AirTemp_Avg_Secondary, AirTemp_Min_Secondary, AirTemp_Min_Time_Secondary, AirTemp_Max_Secondary, and AirTemp_Max_Time_Secondary columns.
In Babase 5.7.5, released on 09 Sep 2025, the old warning system was retired and replaced by Isok. Removing and replacing an entire system sounds like a big change that deserves a larger version "bump", but the old system was almost identical to Isok. Tables have slightly different names and are no longer in the Babase schema, but it's otherwise a pretty small change.
Babase 5.8 was released on 18 Dec 2025. In this update, the system for recording nucleic acid concentration was redone. Before this date, NUCACID_CONC_DATA recorded all concentrations in pg/μL in a dedicated column named "Pg_ul". This update dropped that column and added the Quantity and Unit columns. Also, the NUCACID_CONC_UNITS table and convert_conc() function were added to support this change. The NUCACID_CONCS view underwent the same changes as NUCACID_CONC_DATA. The NUCACIDS_W_CONC view had some changes "under the hood", but is functionally unchanged.
In Babase 5.8.1, released on DD MMM YYYY (not yet), the weather data added the ability for its temperatures to be curated. This included the following changes:
The AirTemp_Min_Curated and AirTemp_Max_Curated columns were added to DIGITAL_WEATHER.
The CURATED_TEMPS view was created.
Babase 4.0 was released on December 18, 2019. In this update, several changes and additions were made to support tracking an individual's group-of-residency. Better support for group fusions was also added.
The following changes make Babase 4.0 incompatible with prior releases:
The GROUPS.Study_Grp column was changed from a boolean to a date. (This change is likewise made in the GROUPS_HISTORY view.)
GROUPS (and GROUPS_HISTORY) has a new To_group column, used to indicate when a group fuses with one or more other groups to make a new group. The new "fusion product" groups are no longer allowed to have a From_group value.
The GROUPS.Supergroup column no longer exists.
The supergroup() function no longer exists.
There is a new MEMBERS.Supergroup column. To better handle group fusions, "supergroup-ness" is now a property of an individual on a date.
On July 13, 2020, several tables and views were added to allow the recording of inventory data. This includes the following:
The TISSUE_DATA table and TISSUES view, for recording data about tissue samples in the inventory.
The NUCACID_DATA table and NUCACIDS view, for recording data about nucleic acid samples in the inventory.
The NUCACID_CONC_DATA table and the NUCACIDS_W_CONC and NUCACID_CONCS views, for recording data about the concentration of nucleic acid samples.
The UNIQUE_INDIVS and POPULATIONS tables, to record the identities of all the possible individuals whose tissue and/or nucleic acid samples appears in the inventory. These tables facilitate the inclusion of individuals from other populations, in addition to the population already recorded in BIOGRAPH.
Several new support tables for validating columns in the above tables.
On September 11, 2020 the TISSUE_TYPES.Tissue_Type and TISSUE_DATA.Tissue_Type columns were changed from an integer to text. This change made the Tissue_Descr column in the TISSUES, NUCACIDS, and NUCACIDS_W_CONC views no longer necessary, so it was removed from all three of those views.
On December 2, 2020 the NUCACID_TYPES.NucAcid_Type and NUCACID_DATA.NucAcid_Type columns were changed from an integer to text. This change made the NucAcid_Descr column in the NUCACIDS and NUCACIDS_W_CONC views no longer necessary, so it was removed from them.
On December 8, 2020 the Exact_Date column was added to INTERACT_DATA and related views. This addition made the special requirement that groomings before 2006-07-01 be recorded with the first day of the month redundant, so this special case was removed.
On September 24, 2021 the WSTATIONS table was updated with the addition of the XYLoc and Loc_Source columns.
On October 26, 2021, several tables and views were added to allow the recording of hormone data. A new function was also created. These additions include the following:
The HORMONE_SAMPLE_DATA table and HORMONE_SAMPLES view, for recording hormone-related data about tissue samples used for hormone analysis.
The HORMONE_PREP_DATA table and HORMONE_PREPS views, for recording the laboratory procedures performed in preparation for a hormone assay.
The HORMONE_RESULT_DATA table and HORMONE_RESULTS views, for recording the raw result of each hormone assay performed on a sample.
The HORMONE_PREP_SERIES table, which facilitates the "many-to-many" relationship between HORMONE_PREP_DATA and HORMONE_RESULT_DATA.
The HORMONE_KITS table, which indicates how the raw values from different hormone assays should be corrected and standardized.
The ESTROGENS, GLUCOCORTICOIDS, PROGESTERONES, TESTOSTERONES, and THYROID_HORMONES views, which show all the assay results for a specific hormone with all of the relevant data about the tissue sample and laboratory procedures performed in preparation for the assay.
The corrected_hormone() function, which uses corrections like those recorded in HORMONE_KITS to calculate the "corrected" concentration of a hormone.
The TISSUES_HORMONES view, for the simultaneous addition of new tissue samples to both TISSUE_DATA and HORMONE_SAMPLE_DATA.
On January 19, 2022, the TEMPMAXS.Unadjusted_Tempmax column was added. To allow the possibility for "adjusted" values in the Tempmax column, constraints requiring it and Tempmin to be multiples of 0.5 were removed.
Babase 3.0 was released on August 1, 2012.
The following changes make Babase 3.0 incompatible with prior releases:
The name of the BIOGRAPH.Dcausestatus column was changed to Dcauseconfidence.
The name of the DISPERSEDATES.Disstatus column was changed to Dispconfidence.
On August 30, 2012 the DSAMPLES.Hairlength column name
was changed to Hairsamples. The datatype was changed from
BOOLEAN to allow numeric values between
0 through 2,
inclusive.
On January 5, 2017 the new columns DcauseNatureConfidence and DcauseAgentConfidence were added to BIOGRAPH. These are intended to clarify and replace the BIOGRAPH.Dcauseconfidence column. Backfilling the new columns is a lengthy procedure, so Dcauseconfidence was not removed until March 14, 2018.
On March 30, 2017, the DSAMPLES and DTCULTURES tables were removed, to allow for the addition of the DART_SAMPLES table (and associated support tables) and the DSAMPLES view. There should be some backward compatibility between the old DSAMPLES table and the new DSAMPLES view--care was taken to change as few column names as possible--but there are new columns in the DSAMPLES view, there will certainly also be incompatibility.
A number of changes were made to Babase in the transition from FoxPro (Babase 1.0) to PostgreSQL (Babase 2.0). This appendix attempts to document changes made to data semantics and, on occasion, data values.
By far the most significant change is that the database itself now performs most data validation. A large number of data validation rules were introduced along with this change.
The 2.0 release of Babase also adds documentation where there was none and includes redesign of some Babase components which were added to Babase 1.0 late in its life. Notable is the entire point sample portion of the database, which was not documented in Babase 1.0 and was redesigned for Babase 2.0. The REPSTATS, CYCSTATS, CYCGAPS and related tables are also new to Babase 2.0 as their 1.0 implementations were never completed or documented. Interpolation was also redesigned and extensively documented.
In Babase 1.0 the GROUPS.Permanent was a boolean
(y or n). In Babase 2.0
it changed to a date, or NULL if the group never became a
permanent group.
The GROUPS.Status column was dropped in Babase 2.0. It was originally intended as a way to mark groups which are no longer censused or which ceased to exist for one reason or another because a group split or group coding changed (particularly with respect to the unknown and suchlike groups used at various times). The functionality of this column is more or less subsumed by the GROUPS.Cease_To_Exist column or obviated by the extensive data cleanup which occurred during the transition of Babase 1.0 to Babase 2.0.
The Statdate is now constrained, when the individual is alive, to be the most recent date on which a census located an individual in a group. Although this was true in practice, the 1.0 system did not require it.
This constraint leads directly to another, when the individual is alive and there are no (non-absent) censuses then the individual's Statdate must be the individual's birth date. Because arbitrary Statdates are not allowed, we prevent automatic changes from erasing manually set Statdates.
Addition of MATUREDATES, RANKDATES, CONSORTDATES, and DISPERSEDATES tables
The MATUREDATES, RANKDATES, CONSORTDATES, and
DISPERSEDATES tables of Babase 2.0 were
columns in the Babase 1.0 BIOGRAPH table. Rather than allow
NULL data values, in Babase 2.0 entire rows are simply not
present when there is no data.
Interpolation and MEMBERS
The interpolation procedure changed somewhat. As the interpolation is what creates the MEMBERS table this appendix also describes the changes made to MEMBERS between 1.0 and 2.0.
Individuals have a row in MEMBERS for every day of their lives.[315]
Interpolation now places individuals in the unknown group when individuals' locations cannot be otherwise assigned, for example outside of the 14 day interpolation limit. Formerly, when the individual could not be place in a group on a particular day the individual had no row in MEMBERS on that day.
Individuals are no longer always placed in a group, the group in which they were last censused, on their Statdate and this “location” no longer interpolates.
When first written, the interpolation procedure was designed to work with females, who are unlikely to be absent from their group for more than 28 days. (Twice the 14 day interpolation limit.) By placing an individual in a group on their Statdate, the group in which they were last censused, the females were assured a row in MEMBERS for every day of their lives. Further, analysis was simplified as each of these rows associated the females with their group (even though at the end of their lives they may not have been present in the group.)
The new interpolation procedure does not consider the Statdate in its determination of the individual's group membership on that day, although, as always, when the Statdate is a death date it does stop interpolation.
There is a change in what happens when an individual is censused absent on his birth day. In the new system, if the individual is censused “absent” on his birth interpolation will “override” the absence and place the individual in his Matgrp group in MEMBERS.
In the old system, if the individual is censused “absent” on his birth interpolation will not “override” the absence and place the individual in a group in MEMBERS. As the individual is expected to be somewhere on his birth, it's expected that there be a demography note made for the individual on that date to give the individual a location ' a row in MEMBERS.
MEMBERS.Interp may now be
NULL. The FoxPro system did not have NULL values. In the new system Interp isNULLwhen interpolation does not know where the nearest locating census is. See Pre-Analyzed Data Disturbs InterpolationThe behavior of interpolation on the last census is now documented.
The interpolation procedure changed during the period of use of Babase 1.0, but the changes were not documented. The primary change was that interpolation was altered so that it did not interpolate if there was no subsequent, absent or not, censuses. This prevented (almost) every living individual currently monitored from having a 14 day “tail” of interpolated values following the last entered census -- a “tail” that would disappear the next time the census information was updated.
The structure of the sexual cycle portion of the database was changed. The CYCLES table became CYCPOINTS and the system became responsible for linking together Mdates, Tdates, and Ddates into CYCLES and computing Seq and Series. The SEXSKINS are automatically associated with cycles. The system also computes automatic Mdates. CYCPOINTS.Source was added to allow tracking of automatically added and estimated data. With the addition of CYCPOINTS the PREGS table links directly to Ddates for conception date and Tdates for resumption dates. PREGS.Resume is automatically calculated unless there is a gap in observation. The CYCGAPS and CYCGAPDAYS tables were added. And the CYCSTATS and REPSTATS tables were modified and made useful.
The PCSKINS table was added to Babase 2.0.
For further information please compare the old and new documentation.
The Babase 1.0 JPSAMPS and FPSAMPS were merged and become POINTS and FPOINTS in Babase 2.0. Along with this change all the support tables used by POINT_DATA and FPOINTS were created.
The Datetime columns on JPSAMPS, FPSAMPS, and ALLMISCS were dropped as they become redundant due to the changes in time representation. (See Time Representation below.)
See The All-Occurrences Focal Point Data below for more changes regarding the time data values.
Babase 1.0 represented times as strings of 5 characters, the 3rd character being a colon and the rest numbers. This was due to the lack of a time data type when the system was first implemented.
Babase 2.0 represents all time values as times, generally using a data type having a precision of 1 second. This facilitates the use of the standard library of time and time interval manipulation operators.
Babase 2.0 simplifies its representation of the times collected using the Psion palmtops by dropping the Datetime column in the tables where it was used. The changes affect the POINT_DATA, ALLMISCS, and INTERACT_DATA tables.
When Babase 1.0 loaded Psion palmtop time data into the FPSAMPS (now POINT_DATA), JPSAMPS (now also POINT_DATA), ADLIBS (now ALLMISCS), and INTERACT_DATA tables it truncated the seconds in the time columns (POINT_DATA.Ptime, ALLMISCS.Atime, Start, and Stop), but retained the seconds in the Datetime column. This means that the time columns of Babase 1.0 recorded a time value up to 59.999 seconds earlier than the Datetime column, the actual time recorded on the Psion palmtop. The program which converts the data from Babase 1.0 to Babase 2.0 uses the Datetime column value, so the new system records the actual Psion palmtop time in its time columns, which now contain a different value than the Babase 1.0 time columns. Babase 2.0 time values are up to 59 seconds later than Babase 1.0's time values.
Views are used to transform time related data on POINT_DATA and INTERACT_DATA. Dates are available in Julian format and times are available as seconds past midnight. If necessary the views can be extended to truncate the seconds if there is an ongoing reason to produce time values that are compatible with Babase 1.0.
The names of some support tables were changed between Babase 1.0 and 2.0.
The following support tables were added in Babase 2.0:
New Support Tables
PALMTOPS (subsequently renamed to SAMPLES_COLLECTION_SYSTEMS)
All things DocBook can be found at The DocBook Project.
The basic DocBook reference is DocBook: The Definitive
Guide.[316] While this book describes how to write DocBook, it
does not describe how to generate output or how to vary the
“look”, the term-of-art is style, of the generated
output. A more gentle introduction can be found in Writing
Documentation Using DocBook. Babase uses the Unix xmlto
command, in conjunction with make
to generate various DocBook output formats, to go into further
detail is beyond the scope of this document. However, as altering
the style of the DocBook output is something done rarely it is
useful for the project to have some reference material on-hand as
a guide when needed.
Those who wish to alter the
style of the Babase documentation should start by reading the
Makefile to see how xmlto is invoked.
Follow this with an examination of the style sheet fragments
supplied to xmlto. These files contain XSL, the
Extensible Stylesheet Language, explained in What is
XSL?. To make further sense of this see the reference
material on styling DocBook. This is covered in DocBook XSL: The
Complete Guide, Part
II. Stylesheet options. Additional detail may be found in
XSL
Frequently Asked Questions, and its companion DocBook Frequently Asked
Questions. The FO
Parameter Reference is the comprehensive list of
formatting “customization variables”. The XSL specifications are
available from the W3C, The
World Wide Web Consortium.
An overview of XML and where XSL fits in can be found at XML: The Big Picture.
[316] Be sure to read the edition that describes the version of DocBook you're using. This text was written for DocBook 4.3.
Using the SET CONSTRAINTS statement to change
the timing of constraints can reduce Babase's functionality.
Specifically, it can make it impossible to add a new pregnancy
into the middle of the sequence of a female's existing
pregnancies.
Note
This appendix is excerpted from the PostgreSQL 9.1 documentation chapter titled “Transactions”.
Transactions are a fundamental concept of all database systems. The essential point of a transaction is that it bundles multiple steps into a single, all-or-nothing operation. The intermediate states between the steps are not visible to other concurrent transactions, and if some failure occurs that prevents the transaction from completing, then none of the steps affect the database at all.
Another important property of transactional databases is closely related to the notion of atomic updates: when multiple transactions are running concurrently, each one should not be able to see the incomplete changes made by others.
In PostgreSQL, a transaction is set up by surrounding the
SQL commands of the transaction with BEGIN and
COMMIT commands. So a banking transaction would
actually look like:
BEGIN;
UPDATE accounts SET balance = balance - 100.00
WHERE name = 'Alice';
-- etc etc
COMMIT;
If, partway through the transaction, we decide we do not
want to commit (perhaps we just noticed that Alice's balance went
negative), we can issue the command ROLLBACK instead
of COMMIT, and all our updates so far will be
canceled.
PostgreSQL actually treats every SQL statement as being
executed within a transaction. If you do not issue a
BEGIN command, then each individual statement has an
implicit BEGIN and (if successful)
COMMIT wrapped around it.
For the most part, database integrity checks are built into the system and it should not be possible to put invalid data into the database. However in some cases, whether for reasons of complexity or for some other reason, some database integrity problems are not caught by the system. There are also questionable cases; situations where the acceptability of the data is dependent upon circumstances. In these cases it is useful for the system to provide a warning and allow the user to decide whether a problem really exists. For these reasons, Babase needs a warning system.
Beginning with Babase 5.7.5, Babase's warnings are managed by the PostgreSQL extension, "Isok". For details about how it works and how to use it, see its documentation.
Table of Contents
Occasionally, a need arises to see an earlier version of the data. For example, an analysis from a publication may need to be revisited, and the user may need to use the data as it appeared at the time of publication rather than the data as it appears today. However, for a variety of reasons, reconstructing earlier "versions" of data in Babase can be difficult or even impossible. To address this problem, beginning with Babase 5.0 all tables became "temporal" tables.
A temporal table is one where all inserts, updates, and deletes to the table are recorded with a timestamp, and earlier versions of the data remain accessible for recall. This allows the user to query a table for its data "as of" a specified date, if desired.
Every temporal table in Babase has a Sys_Period column, used to
record when each row of each table was changed in any way. When
a row in a babase table is
updated or deleted, the "old" version is saved in the table's
corresponding "history" table.
When added to the history table, the exclusive upper bound in
the Sys_Period is set to the
current_timestamp. That is, the date and
time of the UPDATE or
DELETE that moved that version of the row
from the table in babase to
the history table in babase_history.
Thus, the data in babase
can be continually updated, but earlier versions remain
available. See the below example.
It should be emphasized that all details provided here are purely fictional. Specific names, dates, and times are used to avoid ambiguity; they do not refer to any real data, events, or personnel.
Suppose that all tables in Babase were populated with many years' worth of data before the Sys_Period column was added to each of them on 08 Sep 2010, at 07:06:05, and that the babase_history schema and its history tables were created just a few seconds later.
Further, suppose there's an individual named TIM.
Long before any tables became temporal, TIM was recorded
in MATUREDATES as having matured "ON"
2003-02-01:
select * from maturedates where sname='TIM';
sname ¦ matured ¦ mstatus ¦ sys_period
−−−−−−+−−−−−−−−−−−−+−−−−−−−−−+−−−−−−−−−−−−−−−−−−−−−−--−
TIM ¦ 2003-02-01 ¦ O ¦ ["2010-09-08 07:06:05",)
Note that the beginning of the Sys_Period is the time that the column was added, not when this row was added to the table many years earlier. The system does not and cannot say anything about changes in a table before we began recording its history.
Having just been created, the MATUREDATES_HISTORY table in babase_history is empty, and will remain empty until any rows in MATUREDATES are updated or deleted.
On 10 Oct 2010, a data manager realized that there was a typo during data entry, and the year of TIM's Matured should actually be 2002. Upon realizing the mistake, at 10:10:10 she updated the MATUREDATES row with the correct date.
select * from maturedates where sname='TIM';
sname ¦ matured ¦ mstatus ¦ sys_period
−−−−−−+−−−−−−−−−−−−+−−−−−−−−−+−−−−−−−−−−−−−−−−−−−−−−−−−
TIM ¦ 2002-02-01 ¦ O ¦ ["2010-10-10 10:10:10",)
The "old" version of the row, in which TIM matured in 2003, is no longer in MATUREDATES. However, it is retained in babase_history for future recall:
SELECT * FROM babase_history.maturedates_history where sname = 'TIM';
sname ¦ matured ¦ mstatus ¦ sys_period
−−−−−−+−−−−−−−−−−−−+−−−−−−−−−+−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−-
TIM ¦ 2003-02-01 ¦ O ¦ ["2010-09-08 07:06:05","2010-10-10 10:10:10")
Note that the Sys_Period for this "old" version now has an (exclusive) end — the time that the row was updated — and that the Sys_Period of the "current" version in MATUREDATES begins (inclusive) at that same time.
A short time after maturing, TIM migrated to a
nonstudy group and observers lost the ability to identify
him. Several years later (November 2011) he returned to a
study group, but observers didn't recognize him. He was
presumed to be a new, never-before-seen male, and was given
a new name: JIM. In February 2012, when this "new
immigrant" was recorded in the database, JIM was recorded as
having matured "BY" the date that he appeared,
2011-11-01:
select * from maturedates where sname='JIM';
sname ¦ matured ¦ mstatus ¦ sys_period
−−−−−−+−−−−−−−−−−−−+−−−−−−−−−+−−−−−−−−−−−−−−−−−−−−−−--−
JIM ¦ 2011-11-01 ¦ B ¦ ["2012-02-22 22:22:22",)
Having just been added to MATUREDATES, "JIM" does not yet have any rows in MATUREDATES_HISTORY.
Years later, genetic analyses showed that TIM and JIM were the same individual. Having two rows in MATUREDATES for a single individual doesn't make sense, so something needs to be corrected:
select * from maturedates where sname in ('JIM', 'TIM');
sname ¦ matured ¦ mstatus ¦ sys_period
−−−−−−+−−−−−−−−−−−−+−−−−−−−−−+−−−−−−−−−−−−−−−−−−−−−−--−
JIM ¦ 2011-11-01 ¦ B ¦ ["2012-02-22 22:22:22",)
TIM ¦ 2002-02-01 ¦ O ¦ ["2010-10-10 10:10:10",)
At this point, MATUREDATES_HISTORY still only has the one row with TIM's old Matured.
SELECT * FROM babase_history.maturedates_history where sname in ('JIM', 'TIM');
sname ¦ matured ¦ mstatus ¦ sys_period
−−−−−−+−−−−−−−−−−−−+−−−−−−−−−+−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−-
TIM ¦ 2003-02-01 ¦ O ¦ ["2010-09-08 07:06:05","2010-10-10 10:10:10")
There are a few different ways to resolve the situation with TIM/JIM. The two most likely options are explored below.
All of JIM's data in Babase could be merged into the data for TIM. In MATUREDATES, TIM's maturity "ON" 2002 is more informative than JIM's maturity "BY" 2011, so JIM's row would simply need to be removed.
Following the 7 Jun 2018 05:03:09 deletion of JIM's row, JIM's and TIM's data in the two tables will look like this:
select * from maturedates where sname in ('JIM', 'TIM');
sname ¦ matured ¦ mstatus ¦ sys_period
−−−−−−+−−−−−−−−−−−−+−−−−−−−−−+−−−−−−−−−−−−−−−−−−−−−−--−
TIM ¦ 2002-02-01 ¦ O ¦ ["2010-10-10 10:10:10",)
SELECT * FROM babase_history.maturedates_history where sname in ('JIM', 'TIM');
sname ¦ matured ¦ mstatus ¦ sys_period
−−−−−−+−−−−−−−−−−−−+−−−−−−−−−+−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−-
TIM ¦ 2003-02-01 ¦ O ¦ ["2010-09-08 07:06:05","2010-10-10 10:10:10")
JIM ¦ 2011-11-01 ¦ B ¦ ["2012-02-22 22:22:22","2018-06-07 05:03:09")
As the more recently-used ID, it may be preferable to keep the name, JIM. All of TIM's data in Babase would thus be merged with the data for JIM. As mentioned above, TIM's maturity "ON" 2002 is more informative than JIM's maturity "BY" 2011, so JIM's row would need to update its Matured and Mstatus to match those of TIM. Also, TIM's row would need to be removed.
Following the 7 Jun 2018 05:03:09 update of JIM's row and deletion of TIM's, JIM's and TIM's data in the two tables will look like this:
select * from maturedates where sname in ('JIM', 'TIM');
sname ¦ matured ¦ mstatus ¦ sys_period
−−−−−−+−−−−−−−−−−−−+−−−−−−−−−+−−−−−−−−−−−−−−−−−−−−−−--−
JIM ¦ 2002-02-01 ¦ O ¦ ["2018-06-07 05:03:09",)
SELECT * FROM babase_history.maturedates_history where sname in ('JIM', 'TIM');
sname ¦ matured ¦ mstatus ¦ sys_period
−−−−−−+−−−−−−−−−−−−+−−−−−−−−−+−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−-
TIM ¦ 2003-02-01 ¦ O ¦ ["2010-09-08 07:06:05","2010-10-10 10:10:10")
TIM ¦ 2002-02-01 ¦ O ¦ ["2010-10-10 10:10:10","2018-06-07 05:03:09")
JIM ¦ 2011-11-01 ¦ B ¦ ["2012-02-22 22:22:22","2018-06-07 05:03:09")
Knowing that every change to the data is recorded with a
timestamp, it is now possible to "go back in time" and query
tables in the database "as of" a specific time. Unfortunately,
PostgreSQL does not include the syntax AS
OF. However, the range operator @>
effectively means the same thing.
Example I.1. Querying "as of" a date
SELECT *
FROM sometable
WHERE sys_period @> '2022-02-22 12:34:56'::timestamptz;
Note that in this example, only the fictional table SOMETABLE is
being selected-from, so the query will only return rows that 1)
were in the table at 2022-02-22 12:34:56, and 2) are still in
the table now. Rows that were in the table on 2022-02-22
12:34:56 but which have since been removed or updated are in
SOMETABLE_HISTORY in the babase_history schema, which is not
selected-from here.
When querying for a table's data "as of" a specific date
in the past, the best practice is to query both the table in
babase and its history
table in babase_history,
simultaneously. The UNION or UNION
ALL operators are ideal for this.
Example I.2. Querying a table's history "as of" a date
WITH sometable_all AS (SELECT * FROM babase.sometable
UNION
SELECT * FROM babase_history.sometable_history)
SELECT *
FROM sometable_all
WHERE sys_period @> '2022-02-22 12:34:56'::timestamptz;
Querying data from a view is more complicated. Views usually represent data from two or more tables that have been joined together somehow, so recreating data from a view will require that the Sys_Period of each component table is accounted-for. The "best" way to do this depends somewhat on the view itself, so a generalized example about "someview" is not provided here.
See below for specific examples using real tables and a real view.
Returning to the story of TIM and JIM in the previous section, suppose a researcher named Dawn published a paper in 2015 that relied on data from a query that she executed at 2015-05-05 05:05:05. In 2015, it was not yet known that TIM and JIM were two names for the same individual, so Dawn's data includes both names and presumes that they are distinct individuals.
Several years later a new researcher, Pam, wants to revisit Dawn's analysis. As a first step, she needs to recreate Dawn's dataset. Pam cannot do this with the data in the babase schema alone; the TIM/JIM misidentification has already been identified and addressed in her time, so only one of those IDs is still present in Pam's "current" data. She needs the data exactly as it was when Dawn collected it.
Dawn's analysis used data from the BIOGRAPH and MATUREDATES tables, and interactions recorded in the ACTOR_ACTEES view. When collecting data to recreate the analysis, the code Pam would use to collect data from the tables is relatively simple:
-- For BIOGRAPH
WITH biograph_all AS (SELECT * FROM babase.biograph
UNION
SELECT * FROM babase_history.biograph_history)
SELECT *
FROM biograph_all
WHERE sys_period @> '2015-05-05 05:05:05'::timestamptz
AND [WHATEVER OTHER CONSTRAINTS DAWN USED];
-- For MATUREDATES
WITH maturedates_all AS (SELECT * FROM babase.maturedates
UNION
SELECT * FROM babase_history.maturedates_history)
SELECT *
FROM maturedates_all
WHERE sys_period @> '2015-05-05 05:05:05'::timestamptz
AND [WHATEVER OTHER CONSTRAINTS DAWN USED];
Knowing that the ACTOR_ACTEES view is a join between INTERACT_DATA, two instances of PARTS, and two subqueries of MEMBERS, Pam accounted for the Sys_Period of each of those tables and recreated Dawn's May 2015 data using:
WITH dawns_time AS -- Declare the date/time once here so it doesn't
-- need to be retyped for every table
(SELECT '2015-05-05 05:05:05'::timestamptz AS this_time)
, interact_data_all AS (SELECT * FROM babase.interact_data
UNION
SELECT * FROM babase_history.interact_data_history)
, dawns_interact_data AS (SELECT *
FROM interact_data_all
WHERE sys_period @> (SELECT this_time FROM dawns_time))
, parts_all AS (SELECT * FROM babase.parts
UNION
SELECT * FROM babase_history.parts_history)
, dawns_parts AS (SELECT *
FROM parts_all
WHERE sys_period @> (SELECT this_time FROM dawns_time))
, members_all AS (SELECT * FROM babase.members
UNION
SELECT * FROM babase_history.members_history)
, dawns_members AS (SELECT *
FROM members_all
WHERE sys_period @> (SELECT this_time FROM dawns_time))
, dawns_actor_actees AS (SELECT dawns_interact_data.iid AS iid
, dawns_interact_data.sid AS sid
, dawns_interact_data.act AS act
, dawns_interact_data.date AS date
, dawns_interact_data.start AS start
, dawns_interact_data.stop AS stop
, dawns_interact_data.observer AS observer
, actor.partid AS actorid
, COALESCE(actor.sname, '998'::CHAR(3)) AS actor
, (SELECT actorms.grp
FROM dawns_members AS actorms
WHERE actorms.sname = actor.sname
AND actorms.date = interact_data.date) AS actor_grp
, actee.partid AS acteeid
, COALESCE(actee.sname, '998'::CHAR(3)) AS actee
, (SELECT acteems.grp
FROM dawns_members AS acteems
WHERE acteems.sname = actee.sname
AND acteems.date = interact_data.date) AS actee_grp
, dawns_interact_data.handwritten AS handwritten
, dawns_interact_data.exact_date AS exact_date
FROM dawns_interact_data
LEFT OUTER JOIN dawns_parts AS actor
ON (actor.iid = dawns_interact_data.iid AND actor.role = 'R')
LEFT OUTER JOIN dawns_parts AS actee
ON (actee.iid = dawns_interact_data.iid AND actee.role = 'E')
SELECT *
FROM dawns_actor_actees
WHERE [WHATEVER CONSTRAINTS DAWN USED];