A Technical Specification Summary
Anne Ndeti Hubbard
DocBook formattingKarl O. Pinc
DocBook formattingMarch 25, 2009
Table of Contents
- Babase Summarized
- The Babase ER Diagrams
- The Babase Views
- The ACTOR_ACTEES View
- The ANESTH_STATS View
- The BODYTEMP_STATS View
- The CENSUS_DEMOG and CENSUS_DEMOG_SORTED Views
- The CHEST_STATS View
- The CROWNRUMP_STATS View
- The CYCLES_SEXSKINS and CYCLES_SEXSKINS_SORTED Views
- The CYCPOINTS_CYCLES and CYCPOINTS_CYCLES_SORTED Views
- The DSAMPLES View
- The DEMOG_CENSUS and DEMOG_CENSUS_SORTED Views
- The DENT_CODES View
- The DENT_SITES View
- The ESTROGENS View
- The GLUCOCORTICOIDS View
- The GROUPS_HISTORY View
- The HORMONE_PREPS View
- The HORMONE_RESULTS View
- The HORMONE_SAMPLES View
- The HUMERUS_STATS View
- The INTERACT and INTERACT_SORTED Views
- The LOCATIONS_FREE View
- The MATERNITIES View
- The MIN_MAXS View
- The MIN_MAXS_SORTED View
- The MPI_EVENTS View
- The MTD_CYCLES View
- The NUCACID_CONCS View
- The NUCACIDS View
- The NUCACIDS_W_CONC View
- The PARENTS View
- The PCV_STATS View
- The POINTS and POINTS_SORTED Views
- The POTENTIAL_DADS View
- The PROPORTIONAL_RANKS View
- The QUADS View
- The SAMPLES_GOFF View
- The SEXSKINS_CYCLES and SEXSKINS_CYCLES_SORTED Views
- The SEXSKINS_REPRO_NOTES View
- The SWERB view
- The SWERB_DATA_XY View
- The SWERB_DEPARTS view
- The SWERB_GW_LOC_DATA_XY View
- The SWERB_GW_LOCS View
- The SWERB_LOC_GPS_XY view
- The SWERB_LOCS view
- The SWERB_UPLOAD view
- The TESTES_ARC_STATS View
- The TESTES_DIAM_STATS View
- The TESTOSTERONES View
- The THYROID_HORMONES View
- The TISSUES View
- The TISSUES_HORMONES View
- The ULNA_STATS View
- The VAGINAL_PH_STATS View
- The WOUNDPATHOLOGIES View
- The WP_DETAILS_AFFECTEDPARTS View
- The WP_HEALS View
- The WP_REPORTS_OBSERVERS View
- Views Which Add Gid To Tables
- The BIRTH_GRP View
- The ENTRYDATE_GRP View
- The STATDATE_GRP View
- The CONSORTDATES_GRP View
- The CYCGAPDAYS_GRP View
- The CYCGAPS_GRP View
- The CYCSTATS_GRP View
- The DARTINGS_GRP View
- The DISPERSEDATES_GRP View
- The MATUREDATES_GRP View
- The MDINTERVALS_GRP View
- The MMINTERVALS_GRP View
- The RANKDATES_GRP View
- The REPSTATS_GRP View
List of Figures
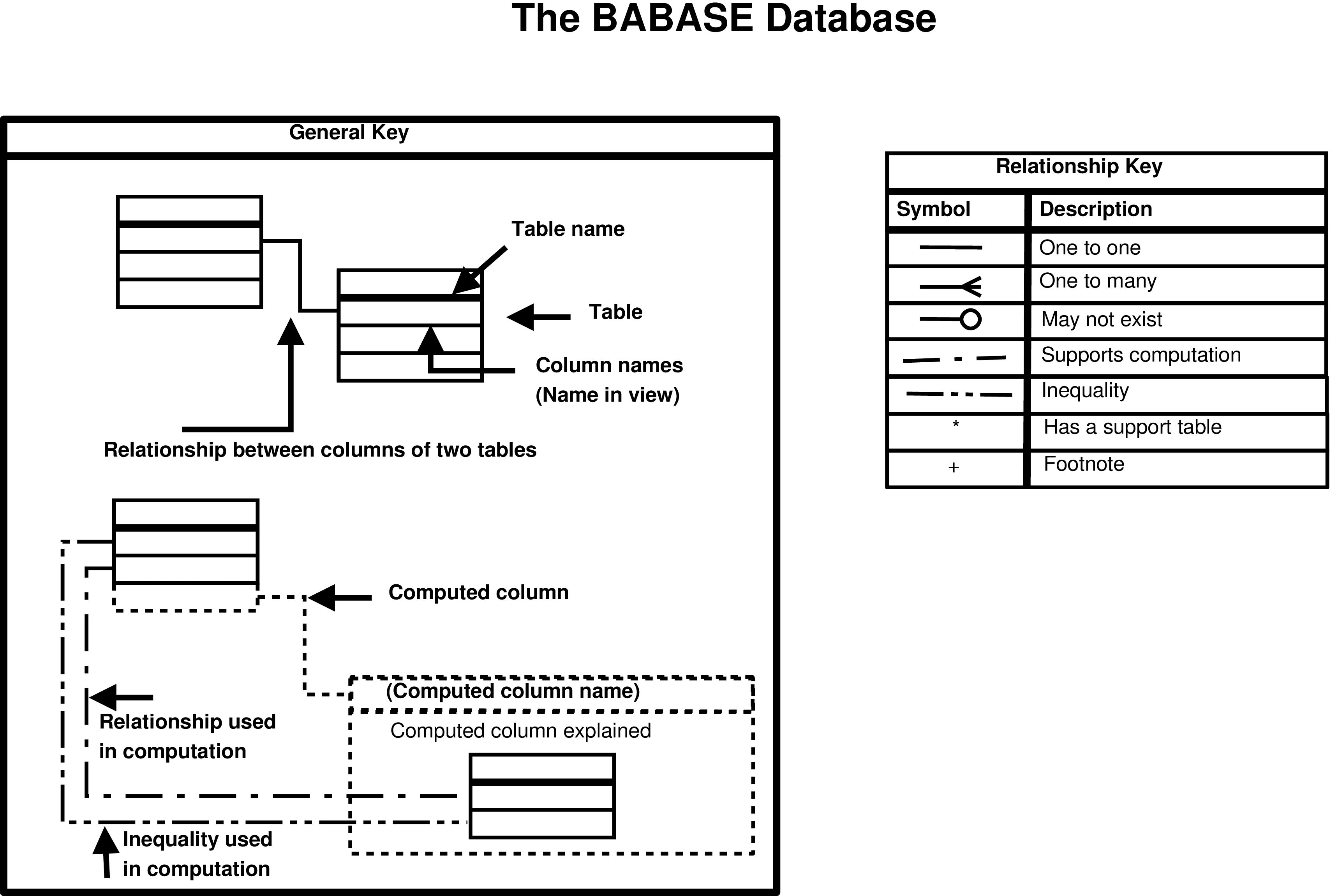
- 1. Key to the Babase Entity Relationship Diagrams
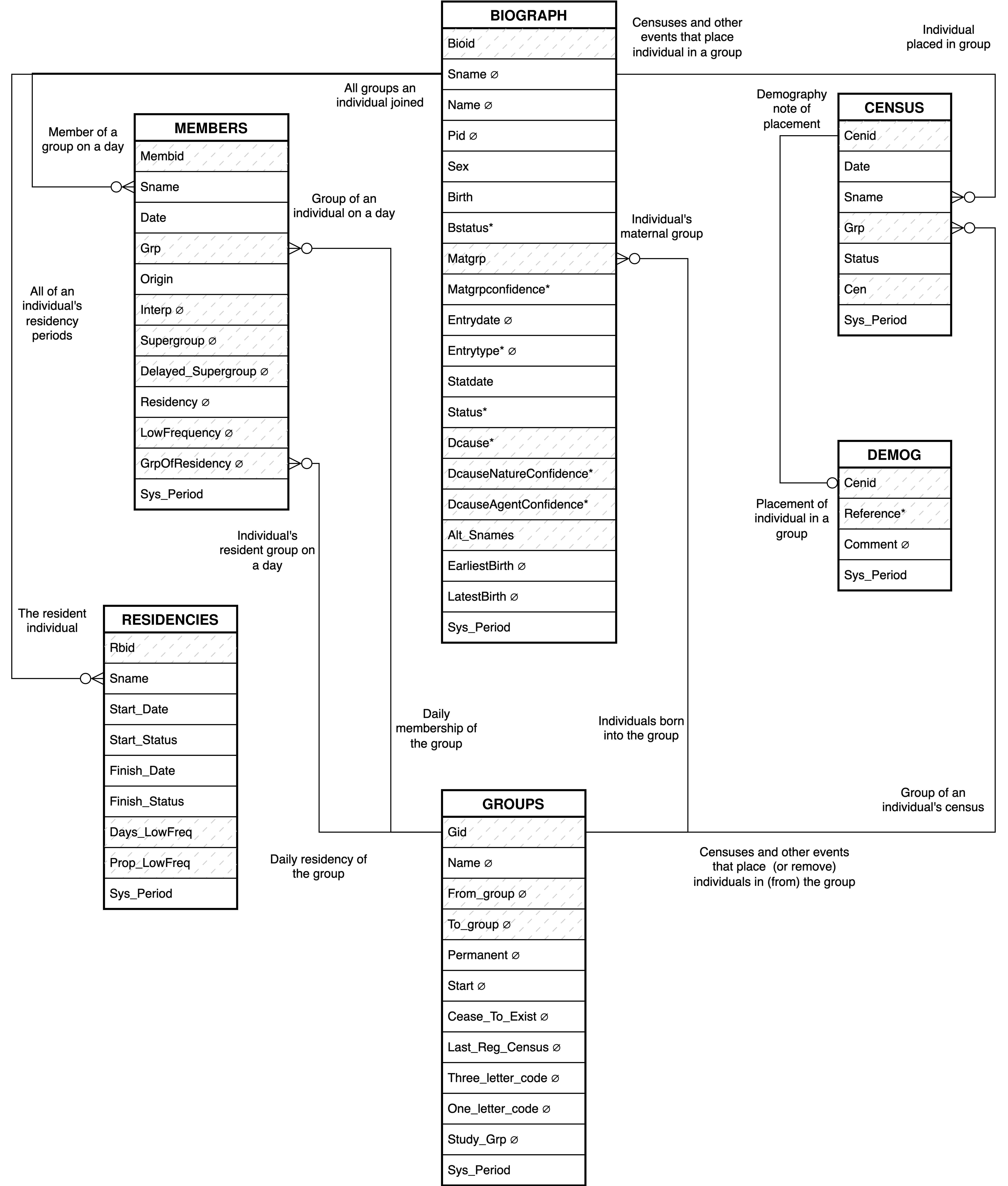
- 2. Babase Group Membership Entity Relationship Diagram
- 3. Babase Life Events Entity Relationship Diagram
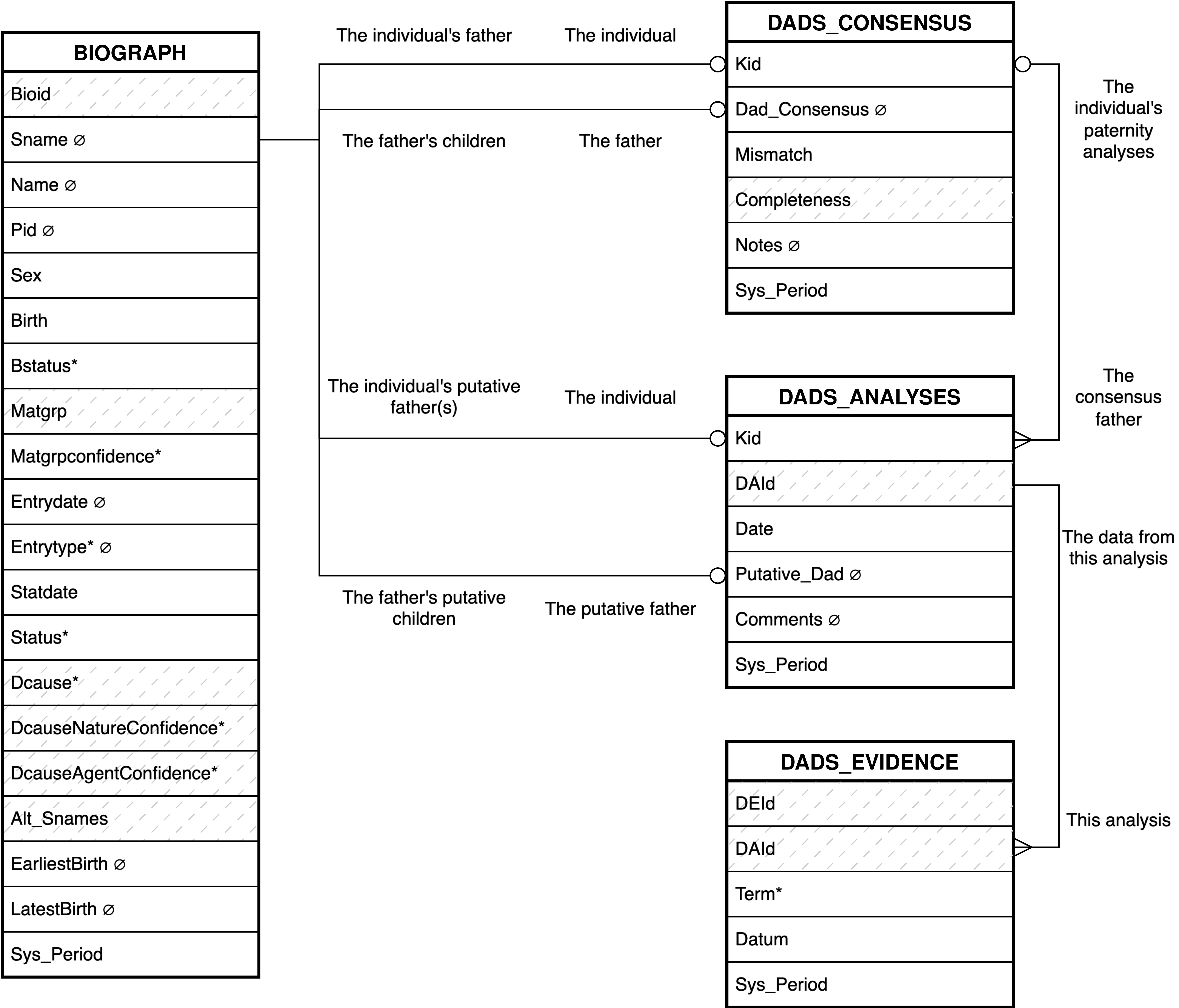
- 4. Babase Paternity Entity Relationship Diagram
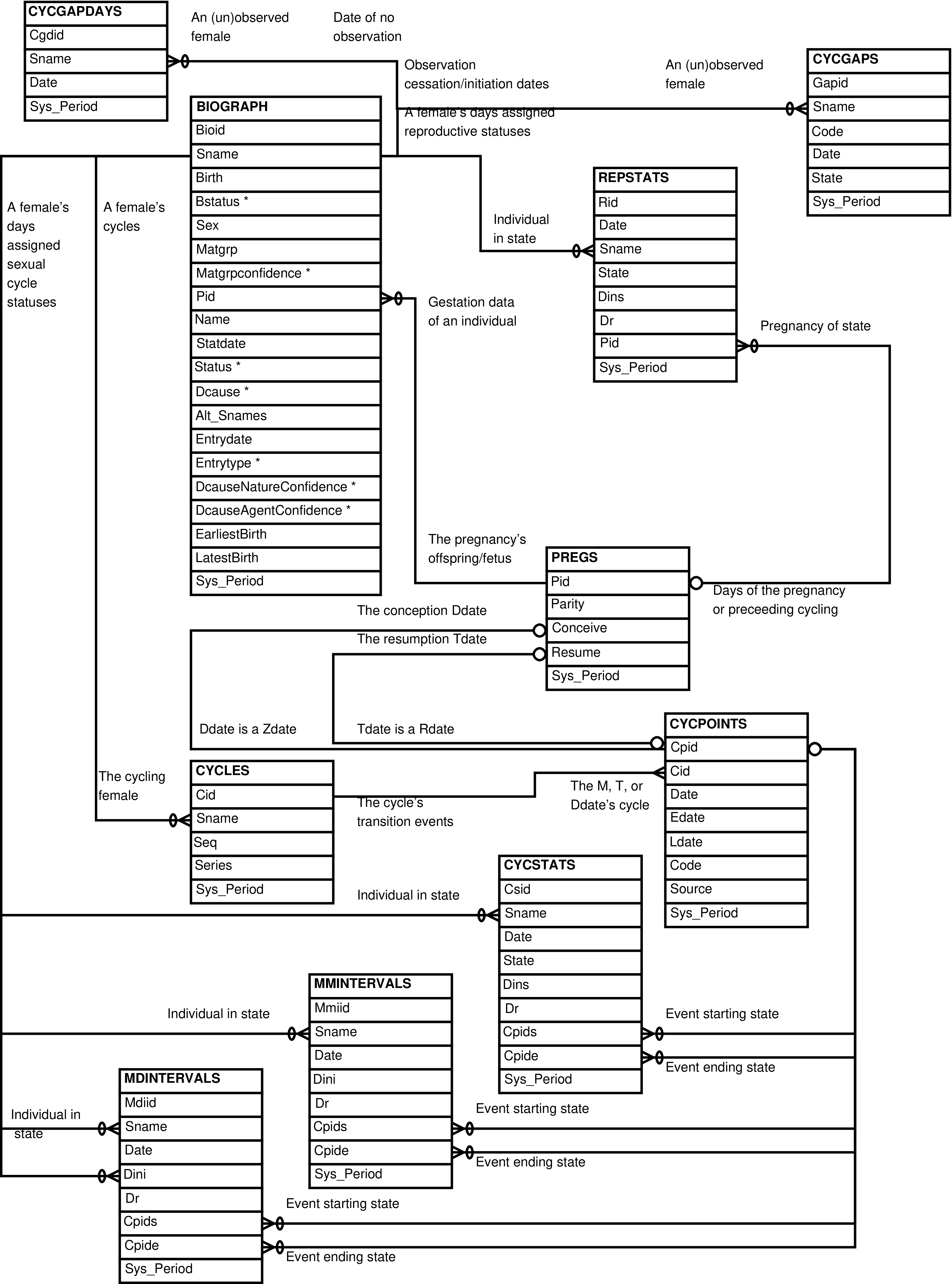
- 5. Babase Sexual Cycle Entity Relationship Diagram
- 6. Babase Sexual Cycle Day-To-Day Tables Entity Relationship Diagram
- 7. Babase Social Interactions Entity Relationship Diagram
- 8. Babase Multiparty Interactions Entity Relationship Diagram
- 9. Babase Darting Logistics and Morphology Entity and Relationship Diagram
- 10. Babase Darting Physiology Entity and Relationship Diagram
- 11. Babase Darting Samples Entity and Relationship Diagram
- 12. Babase Darting Teeth and Ticks Entity and Relationship Diagram
- 13. Babase Inventory Entity Relationship Diagram
- 14. Babase Physical Traits Hormone Data Entity Relationship Diagram
- 15. Babase Physical Traits Hybrid Score Data Entity Relationship Diagram
- 16. Babase Physical Traits Wounds and Pathologies Data Entity Relationship Diagram
- 17. Babase SWERB Core Tables Entity Relationship Diagram
- 18. Babase SWERB Grove/Waterhole Location Tables Entity Relationship Diagram
- 19. Babase Manual Weather Data Entity Relationship Diagram
- 20. Babase Digital Weather Data Entity Relationship Diagram
- 21. Query Defining the ACTOR_ACTEES View
- 22. Entity Relationship Diagram of the ACTOR_ACTEES View
- 23. Query Defining the ANESTH_STATS View
- 24. Entity Relationship Diagram of the ANESTH_STATS View
- 25. Query Defining the BODYTEMP_STATS View
- 26. Entity Relationship Diagram of the BODYTEMP_STATS View
- 27. Query Defining the CENSUS_DEMOG View
- 28. Entity Relationship Diagram of the CENSUS_DEMOG View
- 29. Query Defining the CHEST_STATS View
- 30. Entity Relationship Diagram of the CHEST_STATS View
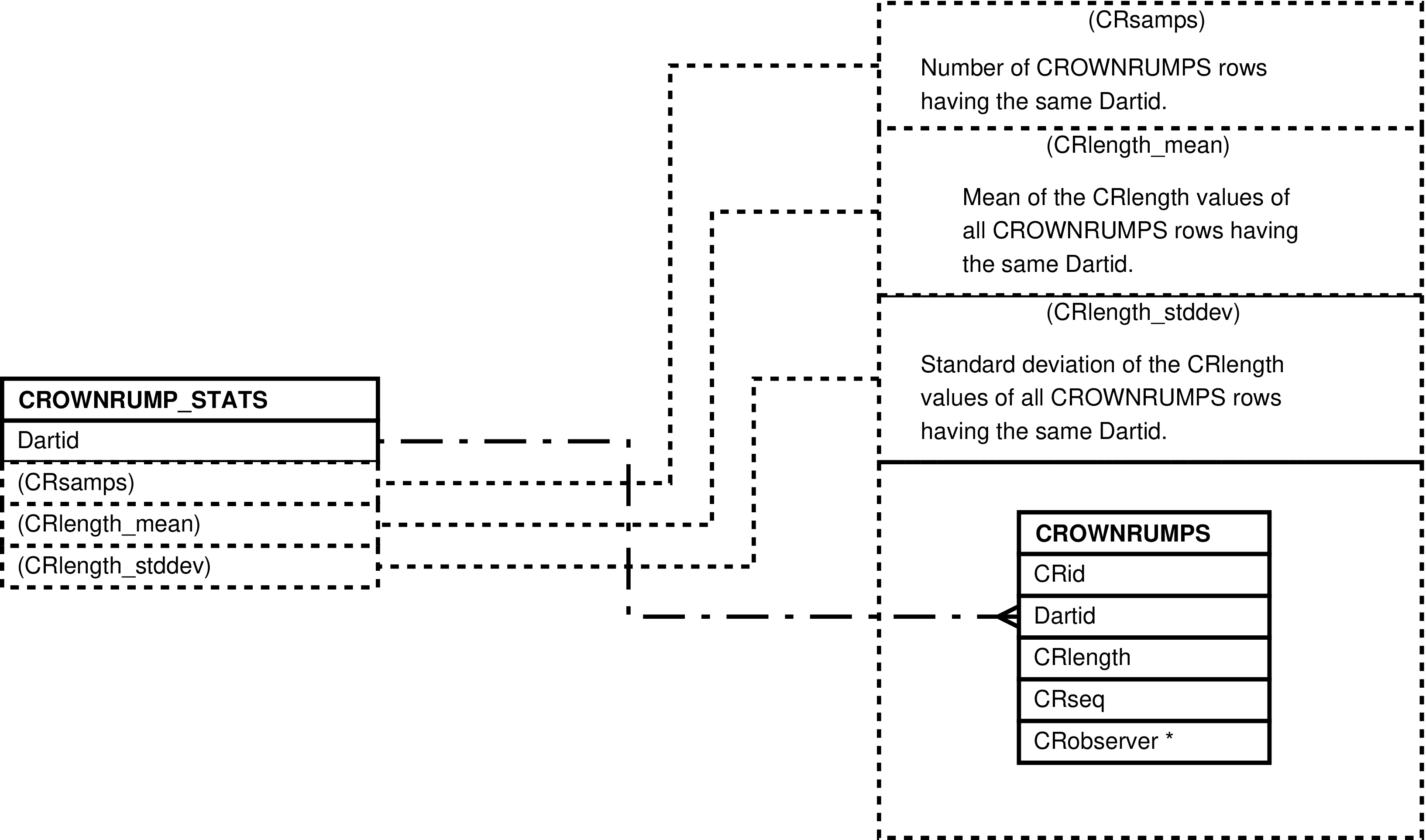
- 31. Query Defining the CROWNRUMP_STATS View
- 32. Entity Relationship Diagram of the CROWNRUMP_STATS View
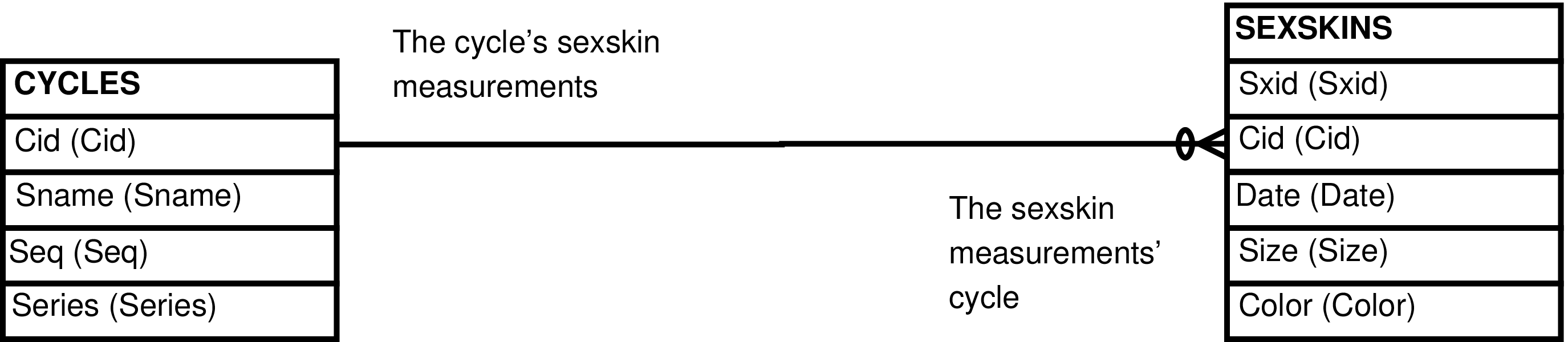
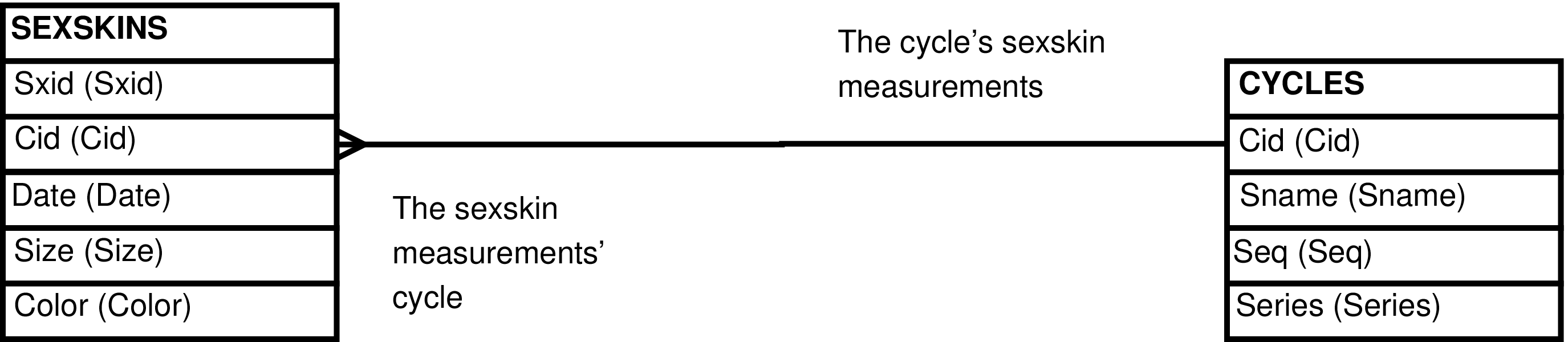
- 33. Query Defining the CYCLES_SEXSKINS View
- 34. Entity Relationship Diagram of the CYCLES_SEXSKINS View
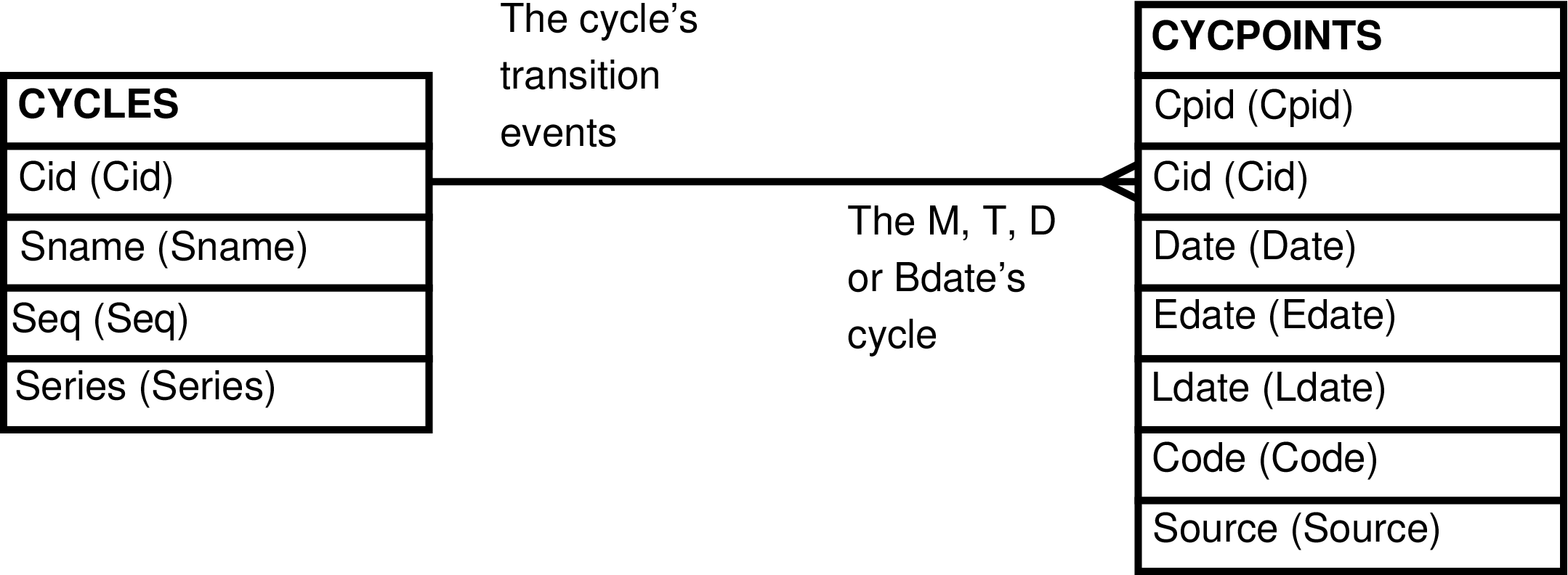
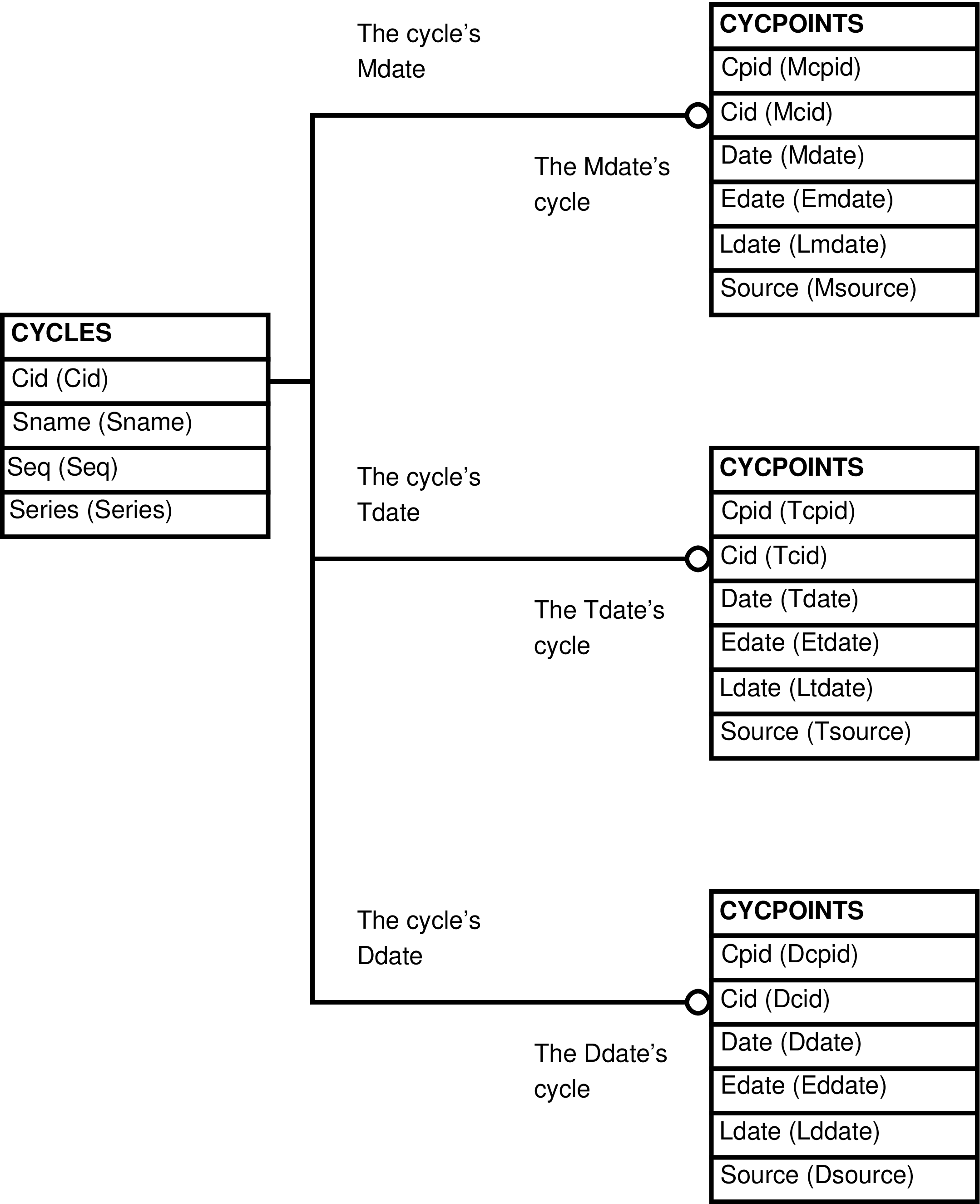
- 35. Query Defining the CYCPOINTS_CYCLES View
- 36. Entity Relationship Diagram of the CYCPOINTS_CYCLES View
- 37. Query Defining the DSAMPLES View
- 38. Query Defining the DEMOG_CENSUS View
- 39. Entity Relationship Diagram of the DEMOG_CENSUS View
- 40. Query Defining the DENT_CODES View
- 41. Entity Relationship Diagram of the DENT_CODES View
- 42. Query Defining the DENT_SITES View
- 43. Entity Relationship Diagram of the DENT_SITES View
- 44. Query Defining the ESTROGENS View
- 45. Entity Relationship Diagram of the ESTROGENS View
- 46. Query Defining the GLUCOCORTICOIDS View
- 47. Entity Relationship Diagram of the GLUCOCORTICOIDS View
- 48. Query Defining the GROUPS_HISTORY View
- 49. Entity Relationship Diagram of the GROUPS_HISTORY View
- 50. Query Defining the HORMONE_PREPS View
- 51. Entity Relationship Diagram of the HORMONE_PREPS View
- 52. Query Defining the HORMONE_RESULTS View
- 53. Entity Relationship Diagram of the HORMONE_RESULTS View
- 54. Query Defining the HORMONE_SAMPLES View
- 55. Entity Relationship Diagram of the HORMONE_SAMPLES View
- 56. Query Defining the HUMERUS_STATS View
- 57. Entity Relationship Diagram of the HUMERUS_STATS View
- 58. Query Defining the INTERACT View
- 59. Entity Relationship Diagram of the INTERACT View
- 60. Query Defining the LOCATIONS_FREE View
- 61. Entity Relationship Diagram of the LOCATIONS_FREE View
- 62. Query Defining the MATERNITIES View
- 63. Entity Relationship Diagram of the MATERNITIES View
- 64. Query Defining the MIN_MAXS View
- 65. Entity Relationship Diagram of the MIN_MAXS View
- 66. Query Defining the MIN_MAXS_SORTED View
- 67. Entity Relationship Diagram of the MIN_MAXS_SORTED View
- 68. Query Defining the MPI_EVENTS View
- 69. Entity Relationship Diagram of the MPI_EVENTS View
- 70. Query Defining the MTD_CYCLES View
- 71. Entity Relationship Diagram of the MTD_CYCLES View
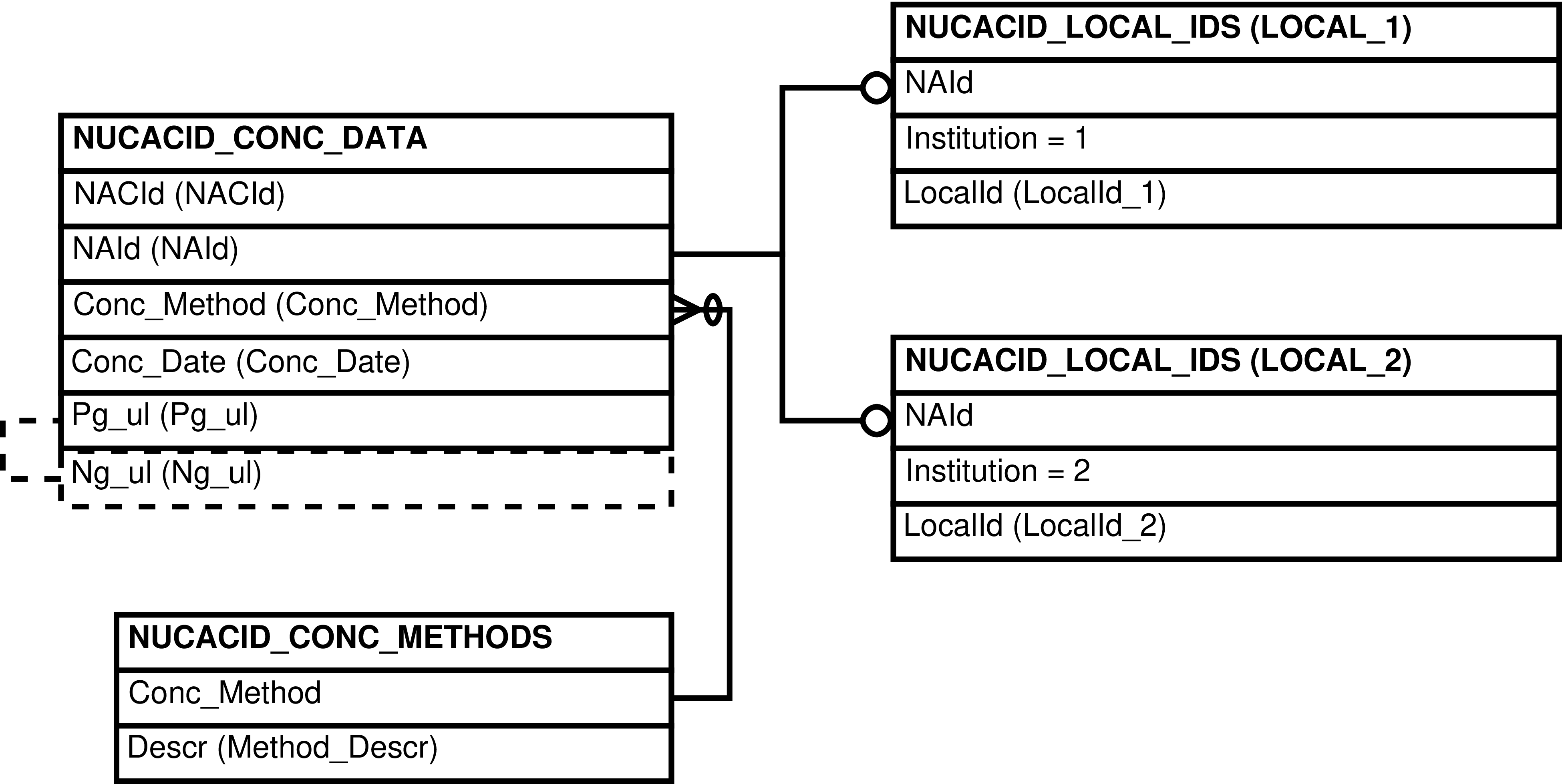
- 72. Query Defining the NUCACID_CONCS View
- 73. Entity Relationship Diagram of the NUCACID_CONCS View
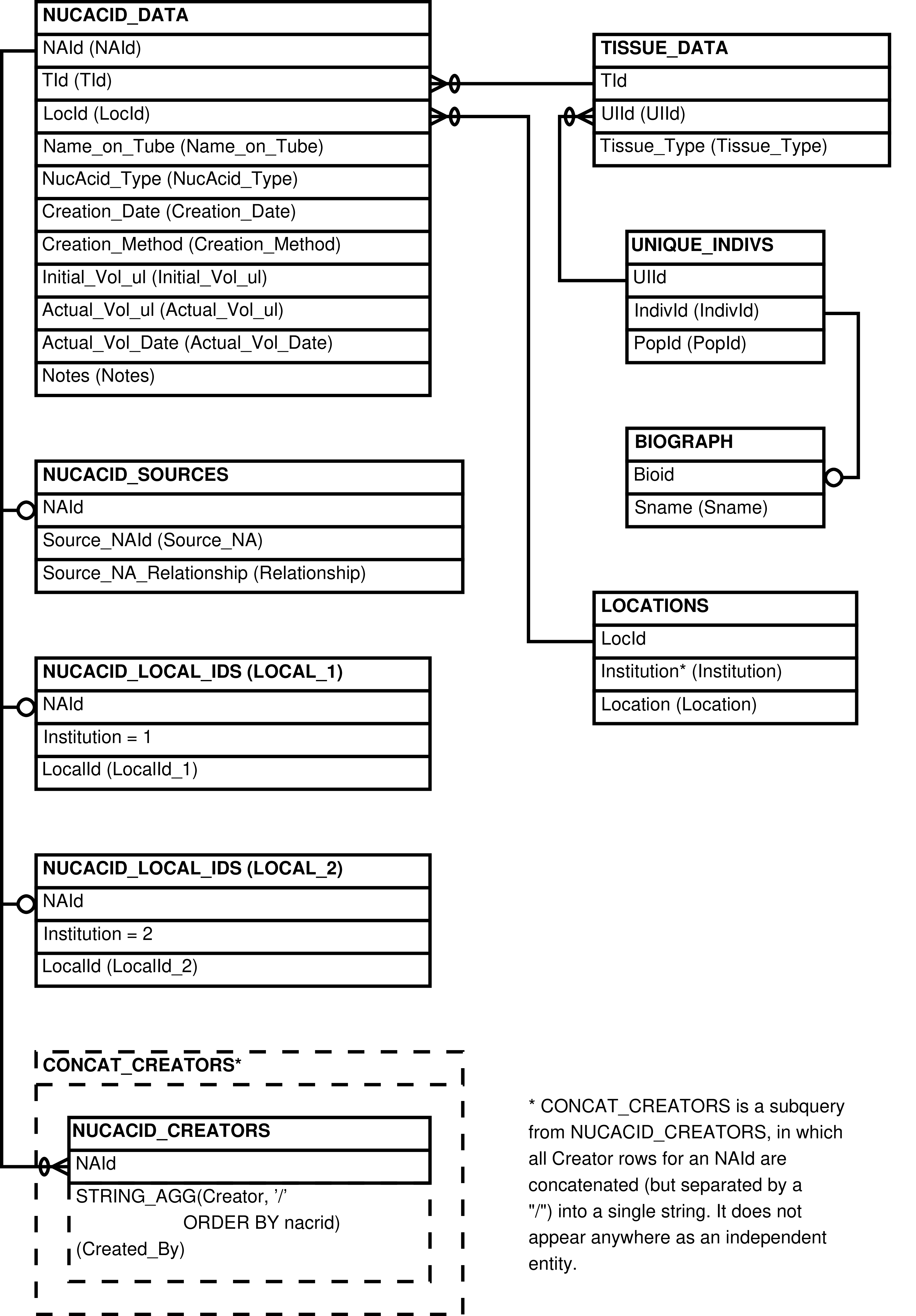
- 74. Query Defining the NUCACIDS View
- 75. Entity Relationship Diagram of the NUCACIDS View
- 76. Query Defining the NUCACIDS_W_CONC View
- 77. Entity Relationship Diagram of the NUCACIDS_W_CONC View
- 78. Query Defining the PARENTS View
- 79. Entity Relationship Diagram of the PARENTS View
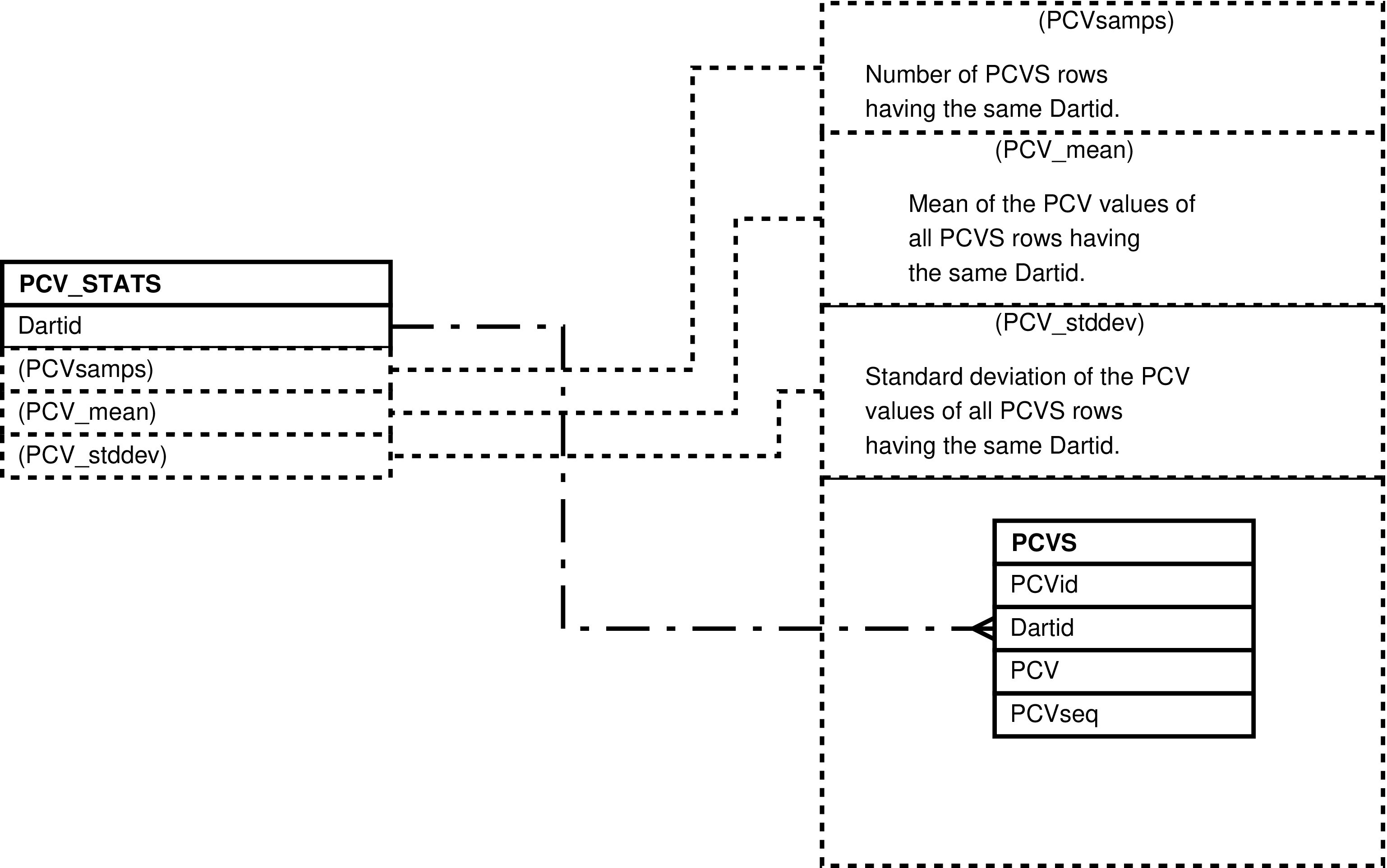
- 80. Query Defining the PCV_STATS View
- 81. Entity Relationship Diagram of the PCV_STATS View
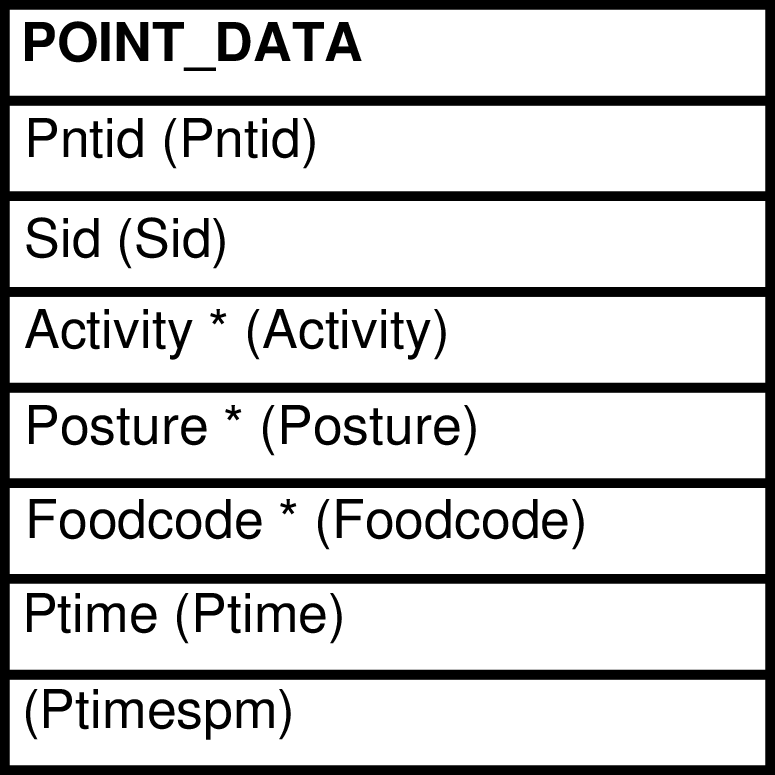
- 82. Query Defining the POINTS View
- 83. Entity Relationship Diagram of the POINTS View
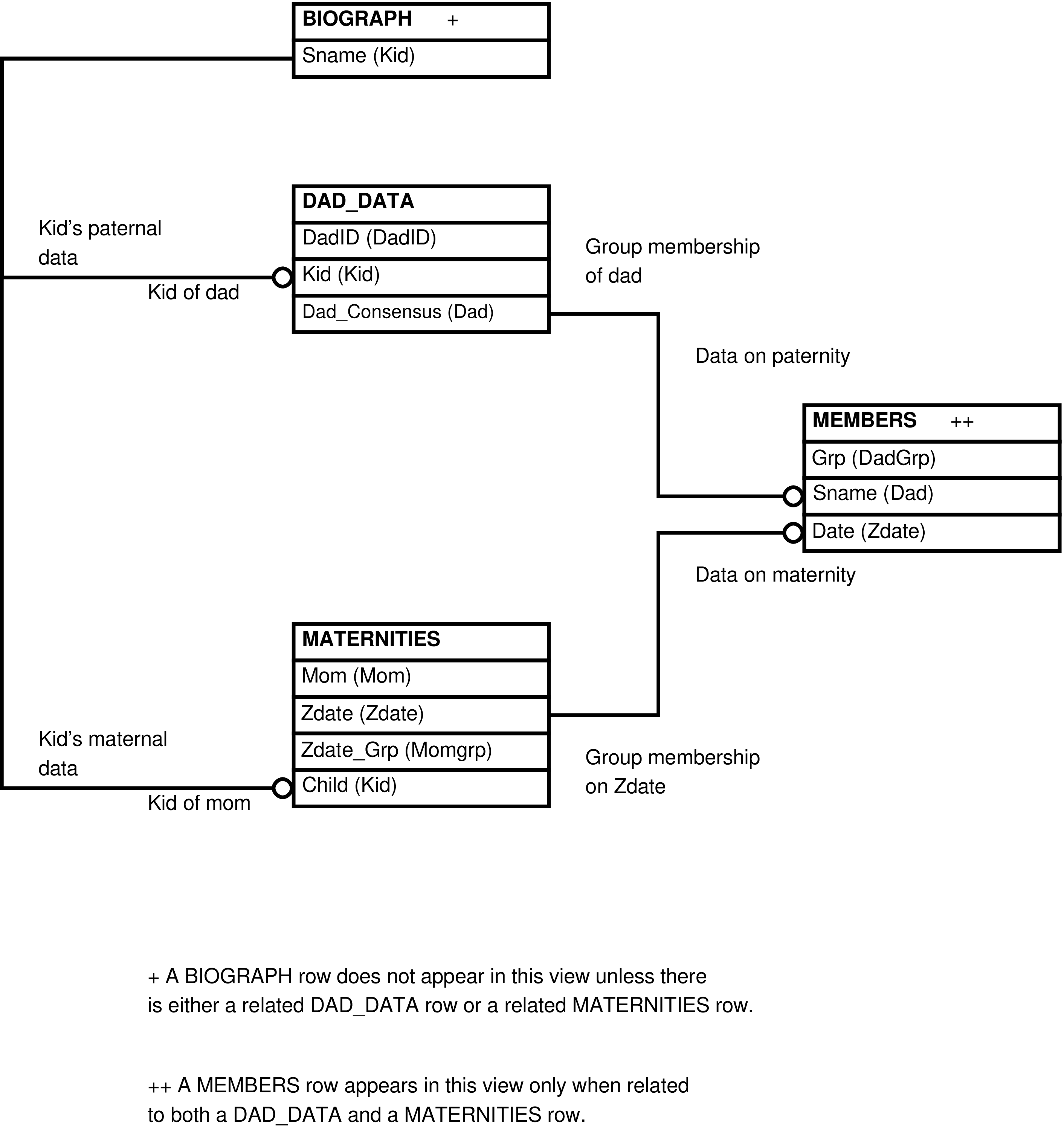
- 84. Query Defining the POTENTIAL_DADS View
- 85. Entity Relationship Diagram of the foundation of the POTENTIAL_DADS View
- 86. Entity Relationship Diagram of that portion of the POTENTIAL_DADS View which places the mother and potential father in the same group during the fertile period
- 87. Entity Relationship Diagram of that portion of the POTENTIAL_DADS View having easily computed columns
- 88. Entity Relationship Diagram of that portion of the POTENTIAL_DADS View involving social interactions
- 89. Query Defining the PROPORTIONAL_RANKS View
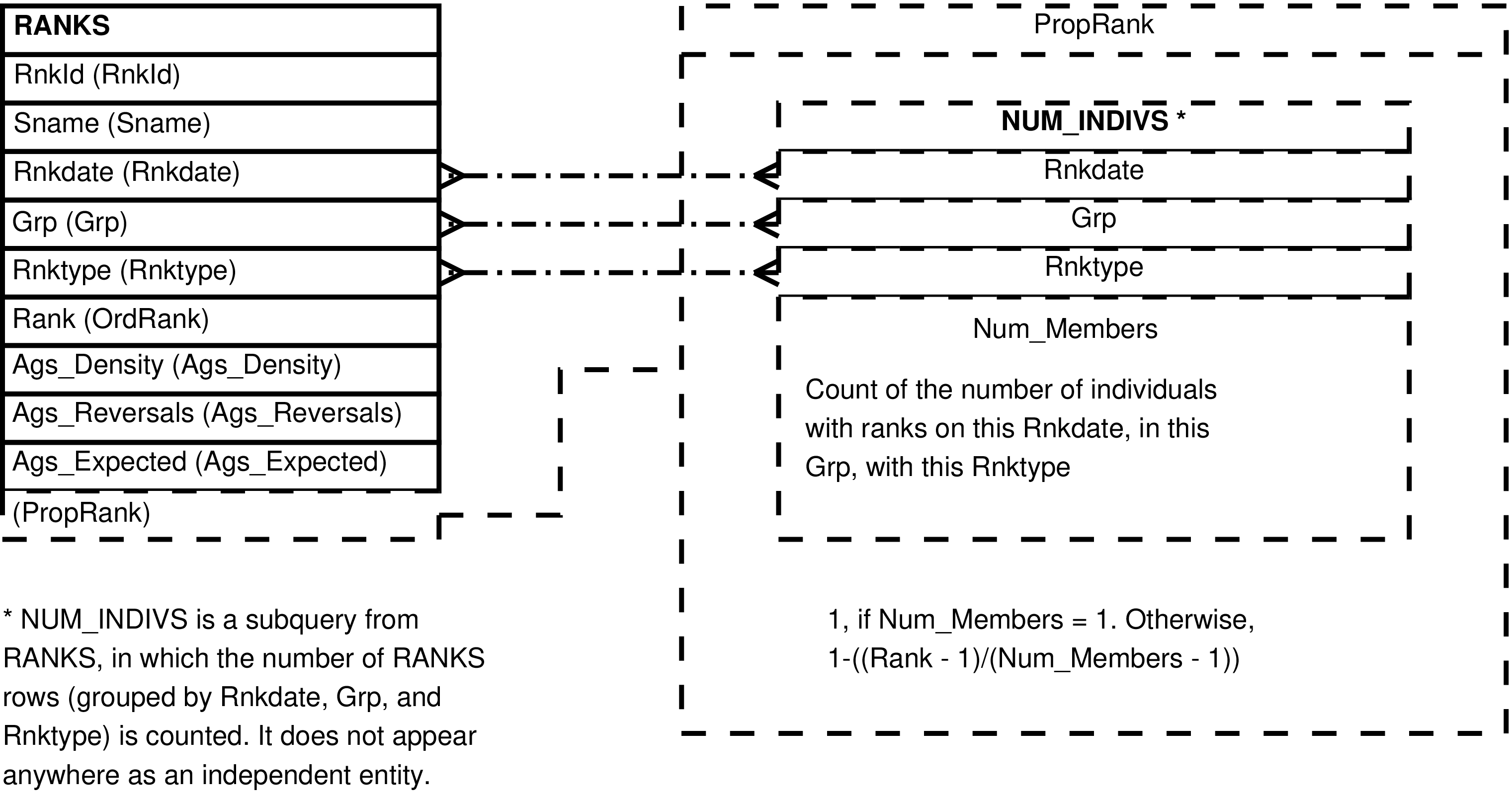
- 90. Entity Relationship Diagram of the PROPORTIONAL_RANKS View
- 91. Query Defining the QUADS View
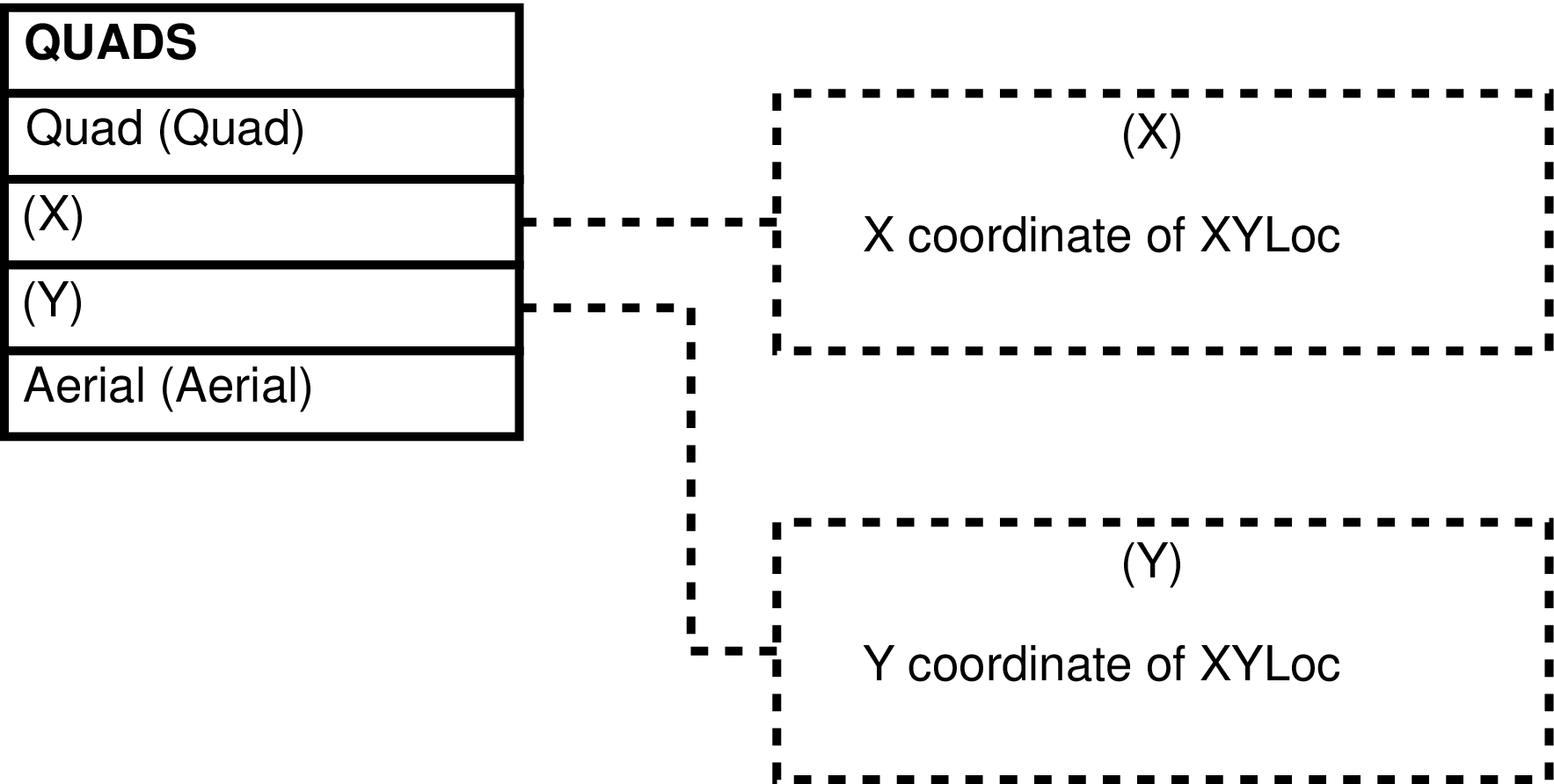
- 92. Entity Relationship Diagram of the QUADS View
- 93. Query Defining the SAMPLES_GOFF View
- 94. Entity Relationship Diagram of the SAMPLES_GOFF View
- 95. Query Defining the SEXSKINS_CYCLES View
- 96. Entity Relationship Diagram of the SEXSKINS_CYCLES View
- 97. Query Defining the SEXSKINS_REPRO_NOTES View
- 98. Entity Relationship Diagram of the SEXSKINS_REPRO_NOTES View
- 99. Query Defining the SWERB View
- 100. Entity Relationship Diagram of the SWERB View
- 101. Query Defining the SWERB_DATA_XY View
- 102. Entity Relationship Diagram of the SWERB_DATA_XY View
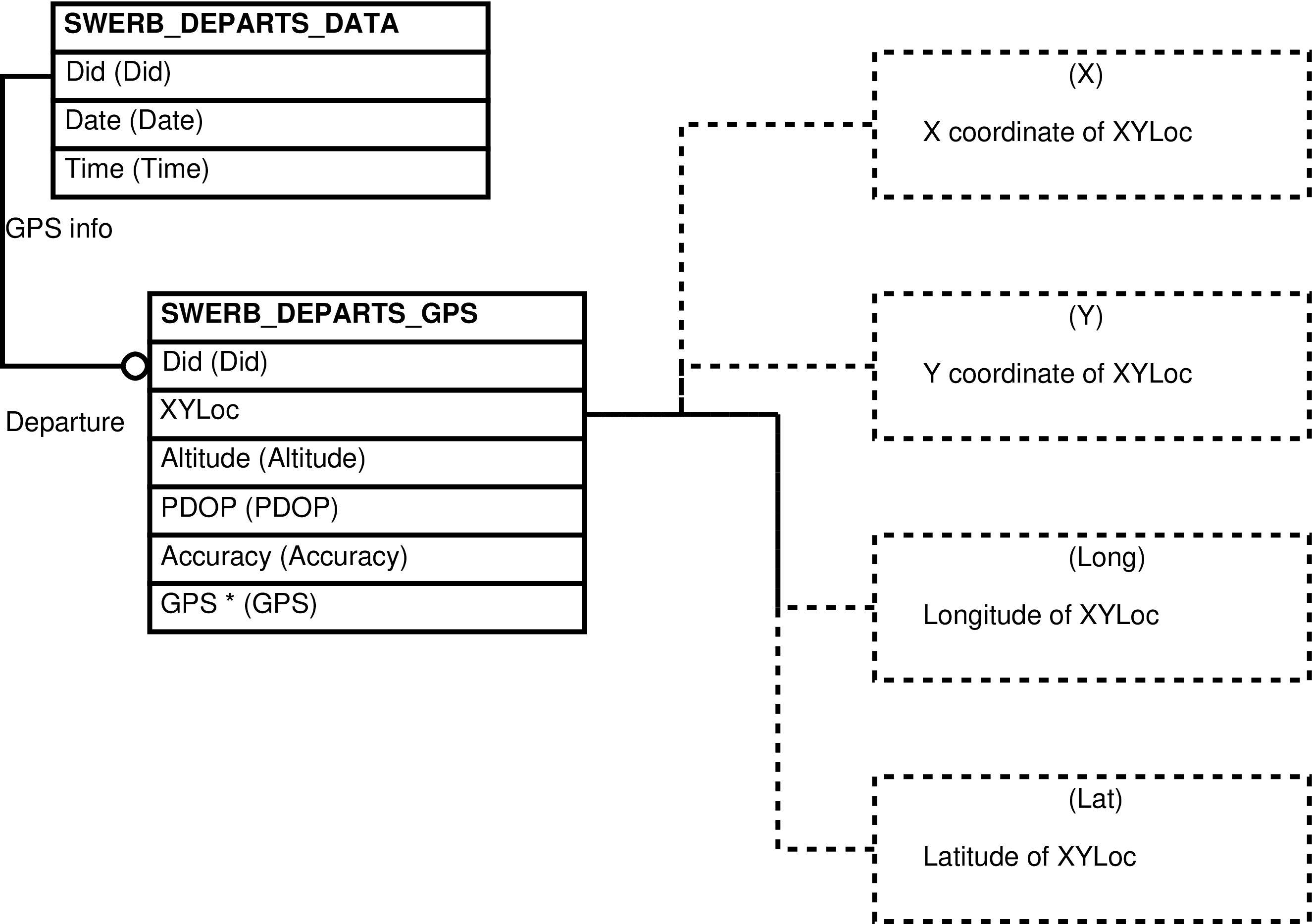
- 103. Query Defining the SWERB_DEPARTS View
- 104. Entity Relationship Diagram of the SWERB_DEPARTS View
- 105. Query Defining the SWERB_GW_LOC_DATA_XY View
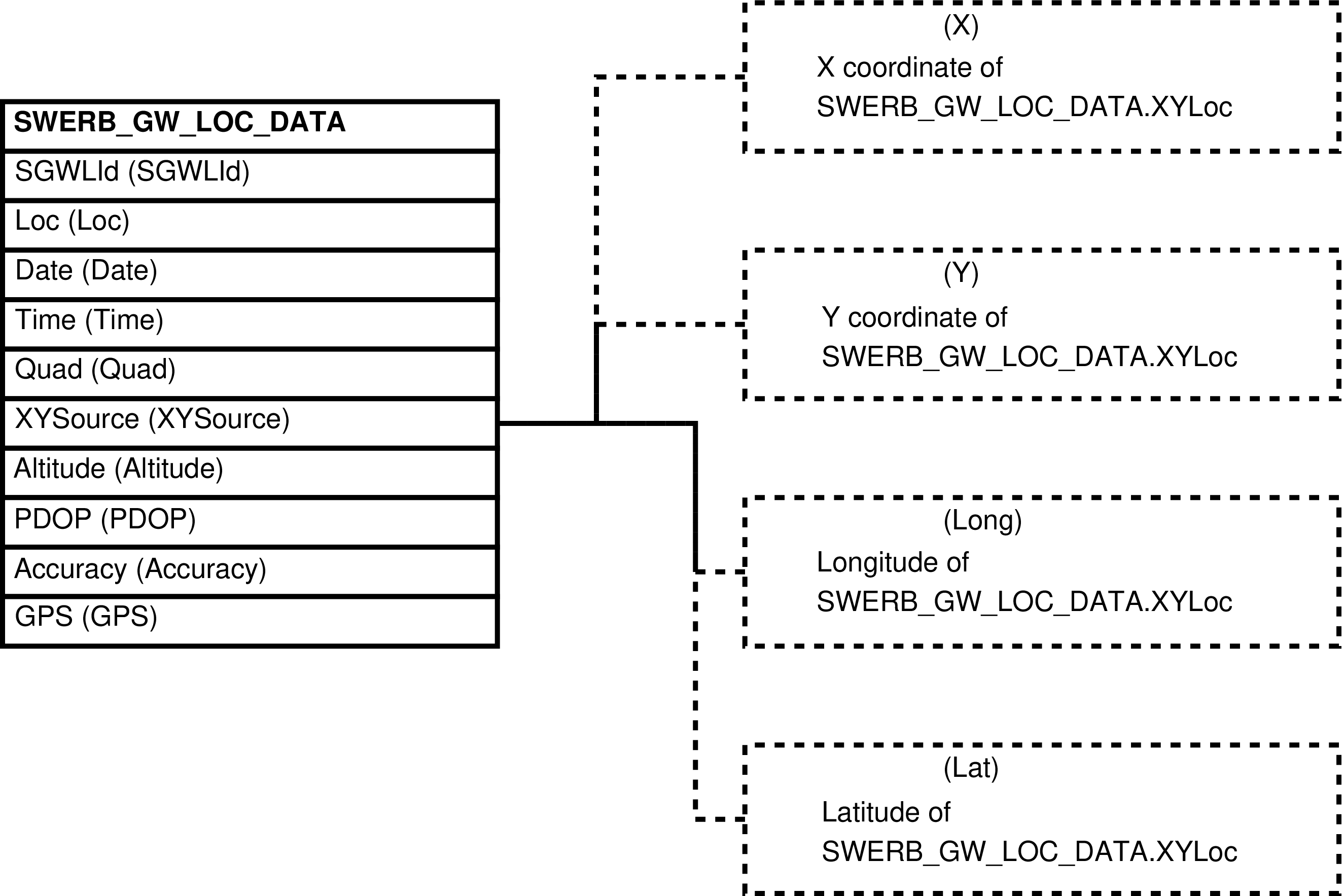
- 106. Entity Relationship Diagram of the SWERB_GW_LOC_DATA_XY View
- 107. Query Defining the SWERB_GW_LOCS View
- 108. Entity Relationship Diagram of the SWERB_GW_LOCS View
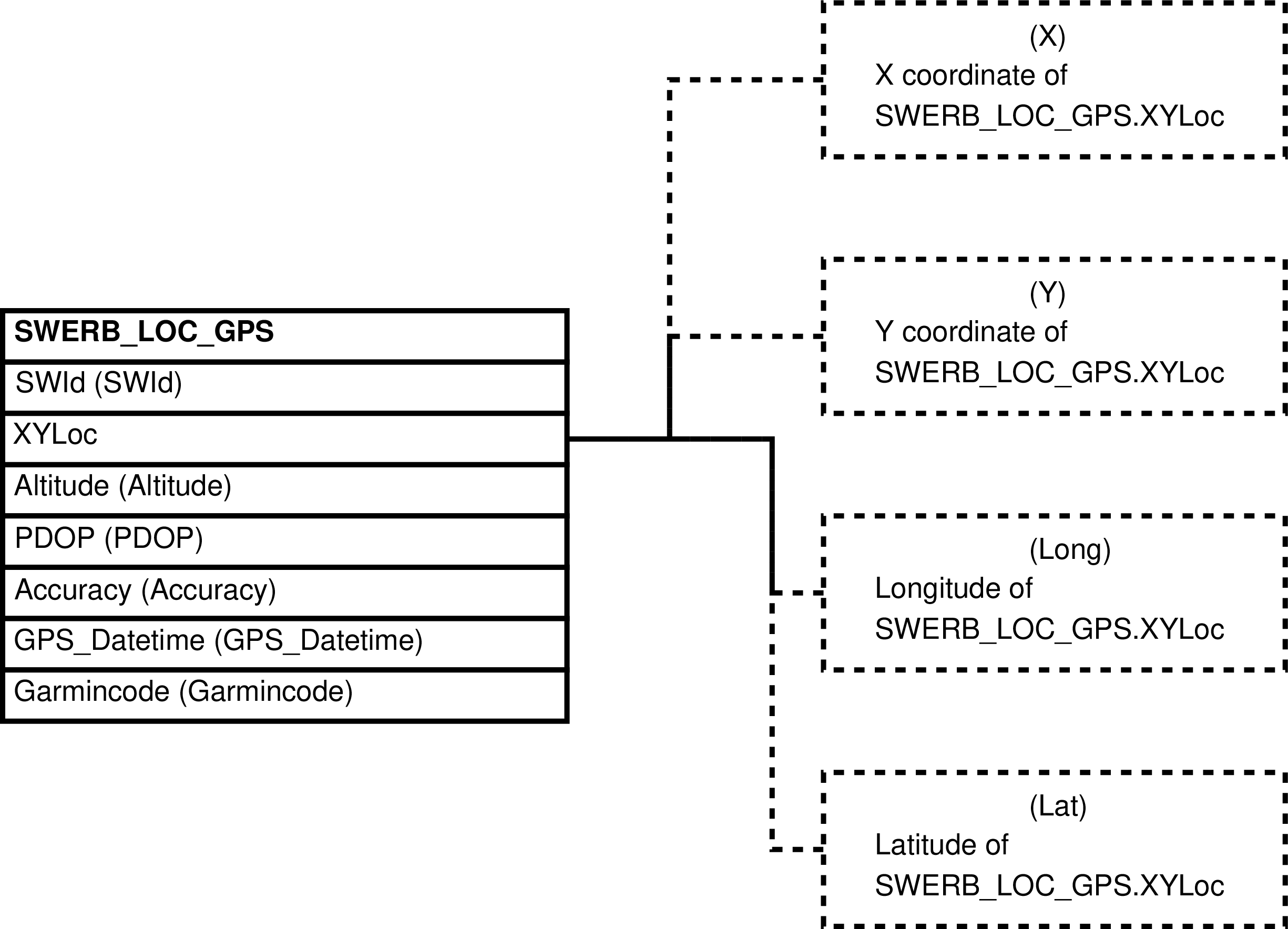
- 109. Query Defining the SWERB_LOC_GPS_XY View
- 110. Entity Relationship Diagram of the SWERB_LOC_GPS_XY View
- 111. Query Defining the SWERB_LOCS View
- 112. Entity Relationship Diagram of the SWERB_LOCS View
- 113. Query Defining the SWERB_UPLOAD View
- 114. Entity Relationship Diagram of the SWERB_UPLOAD View
- 115. Query Defining the TESTES_ARC_STATS View
- 116. Entity Relationship Diagram of the TESTES_ARC_STATS View
- 117. Query Defining the TESTES_DIAM_STATS View
- 118. Entity Relationship Diagram of the TESTES_DIAM_STATS View
- 119. Query Defining the TESTOSTERONES View
- 120. Entity Relationship Diagram of the TESTOSTERONES View
- 121. Query Defining the THYROID_HORMONES View
- 122. Entity Relationship Diagram of the THYROID_HORMONES View
- 123. Query Defining the TISSUES View
- 124. Entity Relationship Diagram of the TISSUES View
- 125. Query Defining the TISSUES_HORMONES View
- 126. Entity Relationship Diagram of the TISSUES_HORMONES View
- 127. Query Defining the ULNA_STATS View
- 128. Entity Relationship Diagram of the ULNA_STATS View
- 129. Query Defining the VAGINAL_PH_STATS View
- 130. Entity Relationship Diagram of the VAGINAL_PH_STATS View
- 131. Query Defining the WOUNDSPATHOLOGIES View
- 132. Entity Relationship Diagram of the WOUNDSPATHOLOGIES View
- 133. Query Defining the WP_DETAILS_AFFECTEDPARTS View
- 134. Entity Relationship Diagram of the WP_DETAILS_AFFECTEDPARTS View
- 135. Query Defining the WP_HEALS View
- 136. Entity Relationship Diagram of the WP_HEALS View, Overall
- 137. Entity Relationship Diagram of the WP_HEALS View for rows with an update to a wound/pathology report
- 138. Entity Relationship Diagram of the WP_HEALS View for rows with an update to a wound/pathology cluster
- 139. Entity Relationship Diagram of the WP_HEALS View for rows with an update to an affected body part
- 140. Query Defining the WP_REPORTS_OBSERVERS View
- 141. Entity Relationship Diagram of the WP_REPORTS_OBSERVERS View
- 142. Query Defining the BIRTH_GRP View
- 143. Entity Relationship Diagram of the BIRTH_GRP View
- 144. Query Defining the ENTRYDATE_GRP View
- 145. Entity Relationship Diagram of the ENTRYDATE_GRP View
- 146. Query Defining the STATDATE_GRP View
- 147. Entity Relationship Diagram of the STATDATE_GRP View
- 148. Query Defining the CONSORTDATES_GRP View
- 149. Entity Relationship Diagram of the CONSORTDATES_GRP View
- 150. Query Defining the CYCGAPDAYS_GRP View
- 151. Entity Relationship Diagram of the CYCGAPDAYS_GRP View
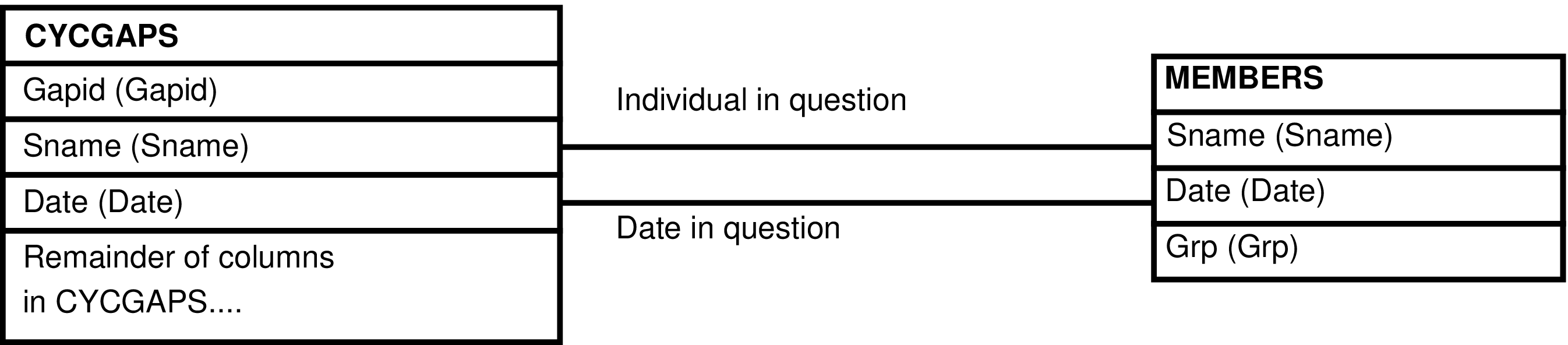
- 152. Query Defining the CYCGAPS_GRP View
- 153. Entity Relationship Diagram of the CYCGAPS_GRP View
- 154. Query Defining the CYCSTATS_GRP View
- 155. Entity Relationship Diagram of the CYCSTATS_GRP View
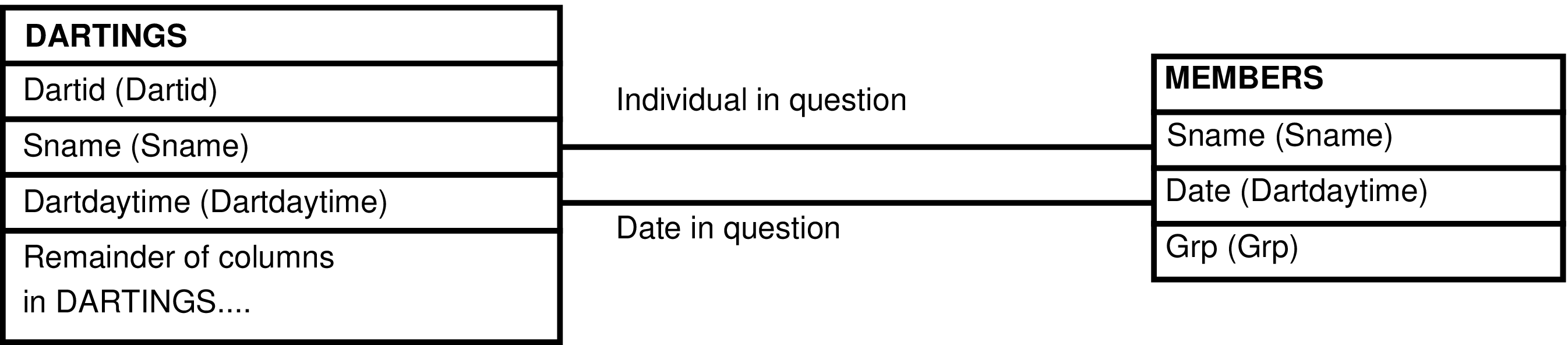
- 156. Query Defining the DARTINGS_GRP View
- 157. Entity Relationship Diagram of the DARTINGS_GRP View
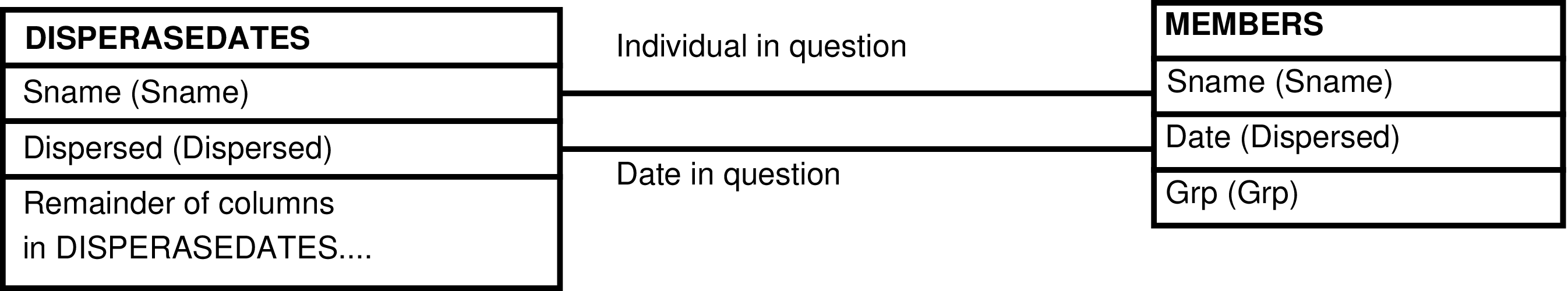
- 158. Query Defining the DISPERSEDATES_GRP View
- 159. Entity Relationship Diagram of the DISPERSEDATES_GRP View
- 160. Query Defining the MATUREDATES_GRP View
- 161. Entity Relationship Diagram of the MATUREDATES_GRP View
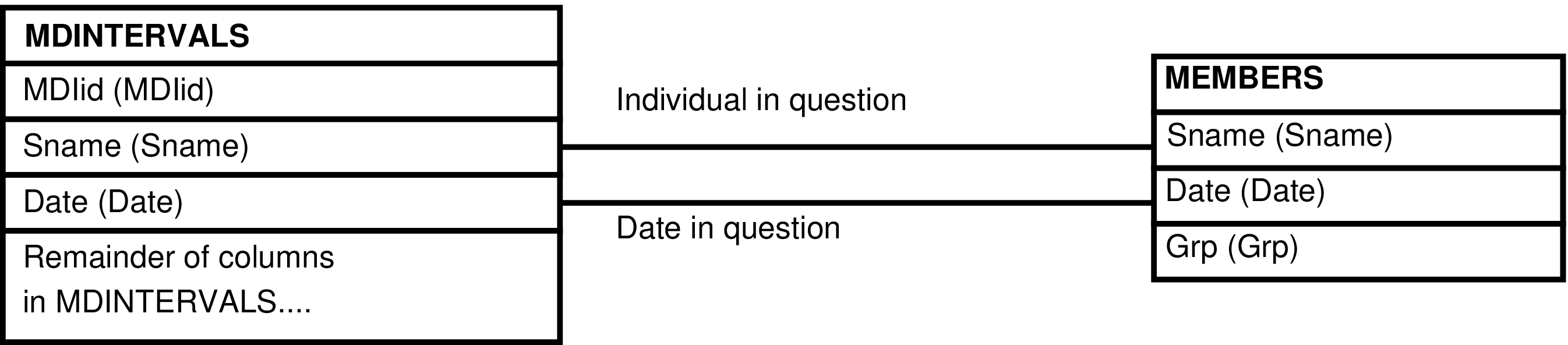
- 162. Query Defining the MDINTERVALS_GRP View
- 163. Entity Relationship Diagram of the MDINTERVALS_GRP View
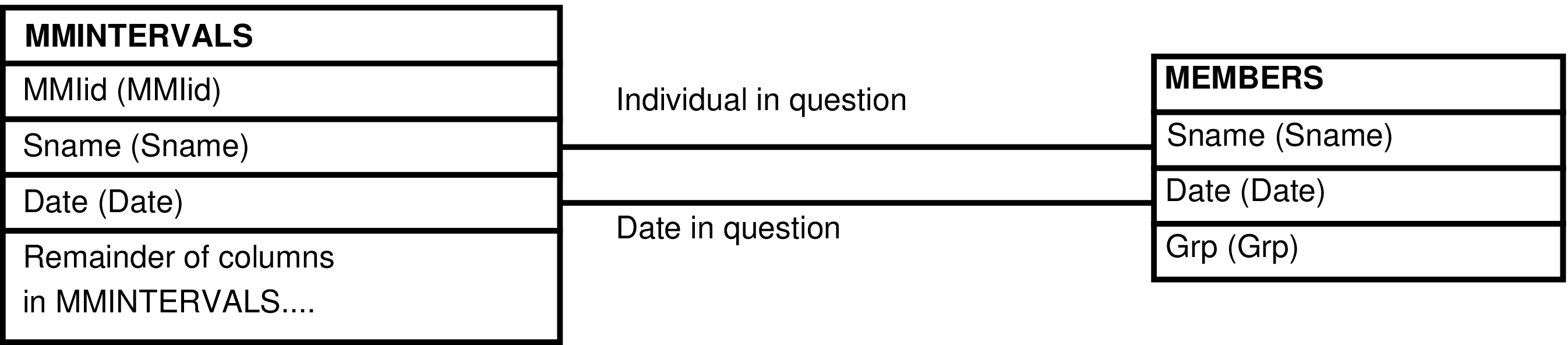
- 164. Query Defining the MMINTERVALS_GRP View
- 165. Entity Relationship Diagram of the MMINTERVALS_GRP View
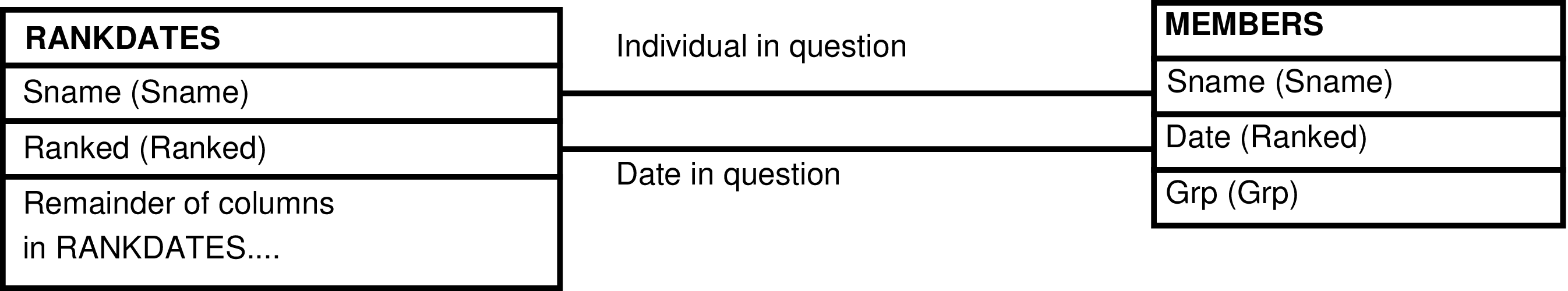
- 166. Query Defining the RANKDATES_GRP View
- 167. Entity Relationship Diagram of the RANKDATES_GRP View
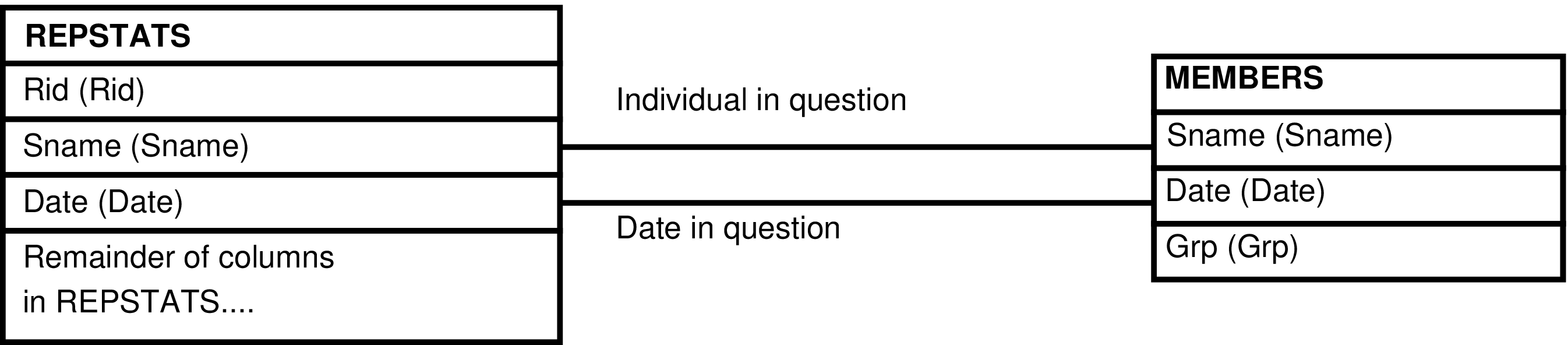
- 168. Query Defining the REPSTATS_GRP View
- 169. Entity Relationship Diagram of the REPSTATS_GRP View
List of Tables
Warning
Tables which have names ending in “_DATA” should not be used, there is always a view of the data in these tables that may be used in their place. Tables ending in “_DATA” may change in future Babase minor releases, breaking queries and programs which use the table. Use of the corresponding views will ensure compatibility with future Babase releases.
Table 1. The Main Babase Tables
| Group Membership and Life Events | |
| Table | One row for each |
| ALTERNATE_SNAMES | rescinded sname |
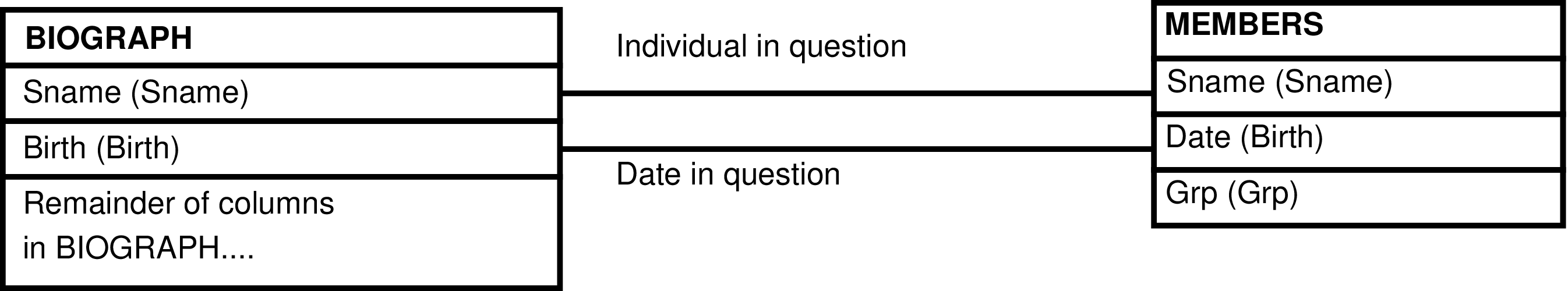
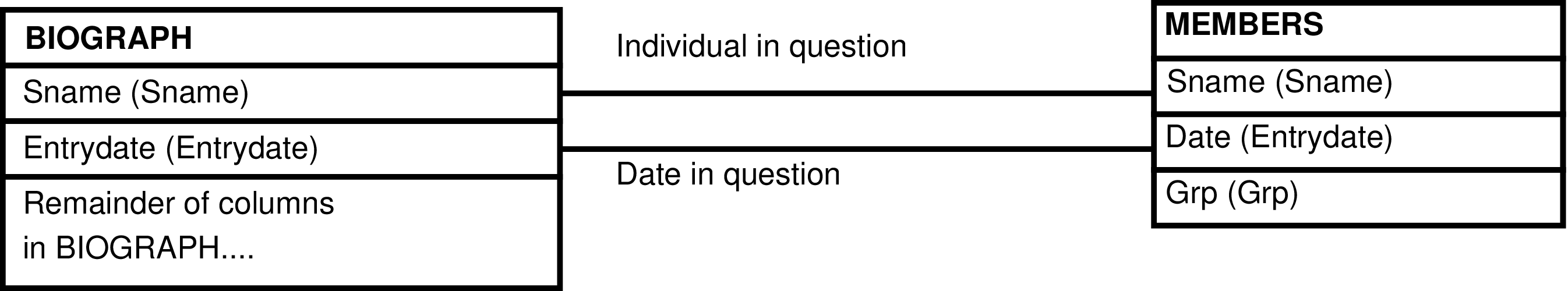
| BIOGRAPH | animal, including fetuses |
| CENSUS | day each individual is (or is not) observed in a group |
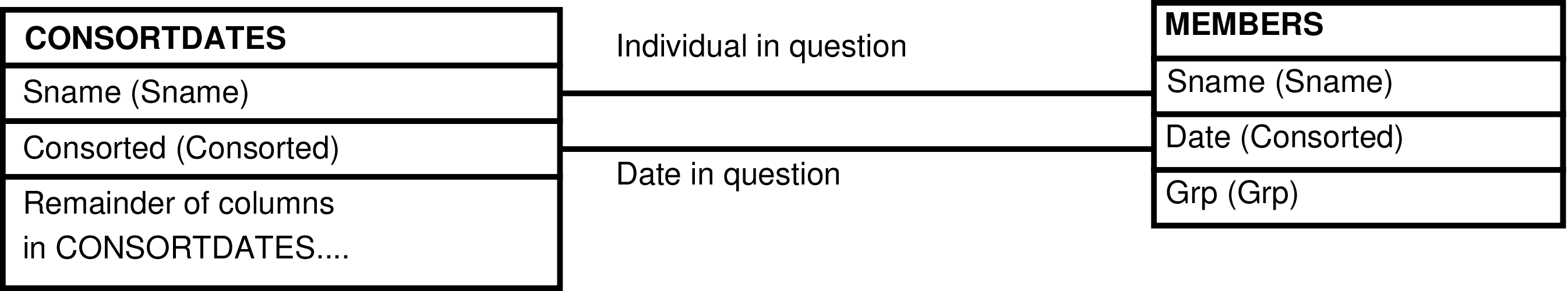
| CONSORTDATES | male who has a known first consortship |
| DEMOG | mention of an individual's presence in a group within a field textual note |
| DISPERSEDATES | male who has left his maternal study group |
| GROUPS | group (including solitary males) |
| MATUREDATES | individual who is sexually mature |
| RANKDATES | individual[a] who has attained adult rank |
| Analyzed: Group Membership and Life Events | |
| Table | One row for each |
| DADS_ANALYSES | paternity analysis |
| DADS_CONSENSUS | kid with a known dad |
| DADS_EVIDENCE | datum used in a paternity analysis |
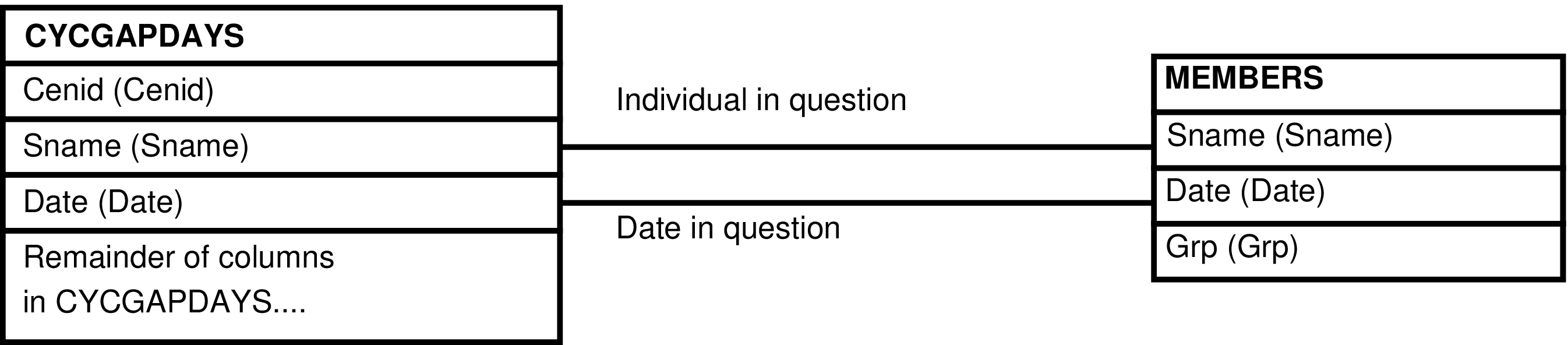
| MEMBERS | day each individual is alive |
| RANKS | month each individual is ranked in each group |
| RESIDENCIES | bout of each individual's residency |
| Physical Traits | |
| Table | One row for each |
| WP_AFFECTEDPARTS | body part affected by a specific wound/pathology |
| WP_DETAILS | wound or pathology cluster indicated on a report |
| WP_HEALUPDATES | update on progress of wound/pathology healing |
| WP_REPORTS | wound/pathology report |
| Analyzed: Physical Traits | |
| Table | One row for each |
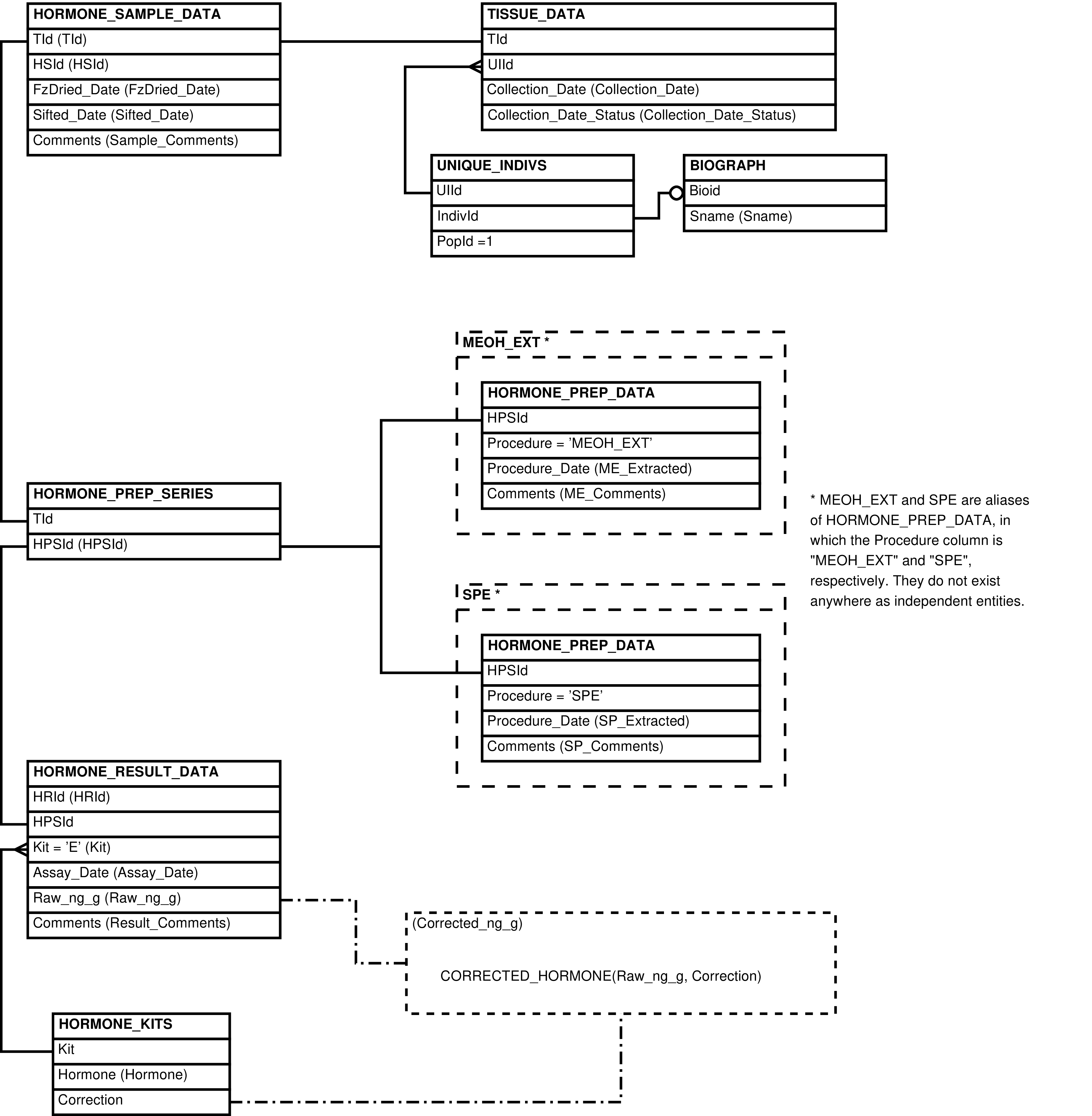
| HORMONE_KITS | kit or protocol used to assay hormone concentration |
| HORMONE_PREP_DATA | laboratory preparation performed on a sample in the specified series |
| HORMONE_PREP_SERIES | series of preparations and assays performed on a sample |
| HORMONE_RESULT_DATA | assay for hormone concentration in a sample |
| HORMONE_SAMPLE_DATA | tissue sample used in hormone analysis |
| HYBRIDGENE_ANALYSES | analysis of genetic hybrid scores |
| HYBRIDGENE_SCORES | genetic hybrid score for an individual from an analysis |
| HYBRIDMORPH_OBSERVERS | observer in a morphological hybrid score report |
| HYBRIDMORPH_REPORTS | morphological hybrid scoring event, per scored individual |
| HYBRIDMORPH_SCORE_DATA | morphological hybrid score for a particular trait |
| Sexual Cycles | |
| Table | One row for each |
| CYCGAPS | female for each initiation or cessation of a continuous period of observation |
| CYCLES | female's cycle (complete or not) |
| CYCPOINTS | Mdate (menses), Tdate (turgesence onset), or Ddate (deturgesence onset) date of each female |
| PREGS | time a female becomes pregnant |
| SEXSKINS | sexskin measurement of each female |
| The Sexual Cycle Day-By-Day Tables | |
| Table | One row for each |
| CYCGAPDAYS | female for each day within a period during which there is not continuous observation |
| CYCSTATS | day each female is cycling -- by M, T and Ddates |
| MDINTERVALS | day each female is cycling and is between M and Ddates |
| MMINTERVALS | day each female is cycling -- by Mdates |
| REPSTATS | day each female has a known reproductive state |
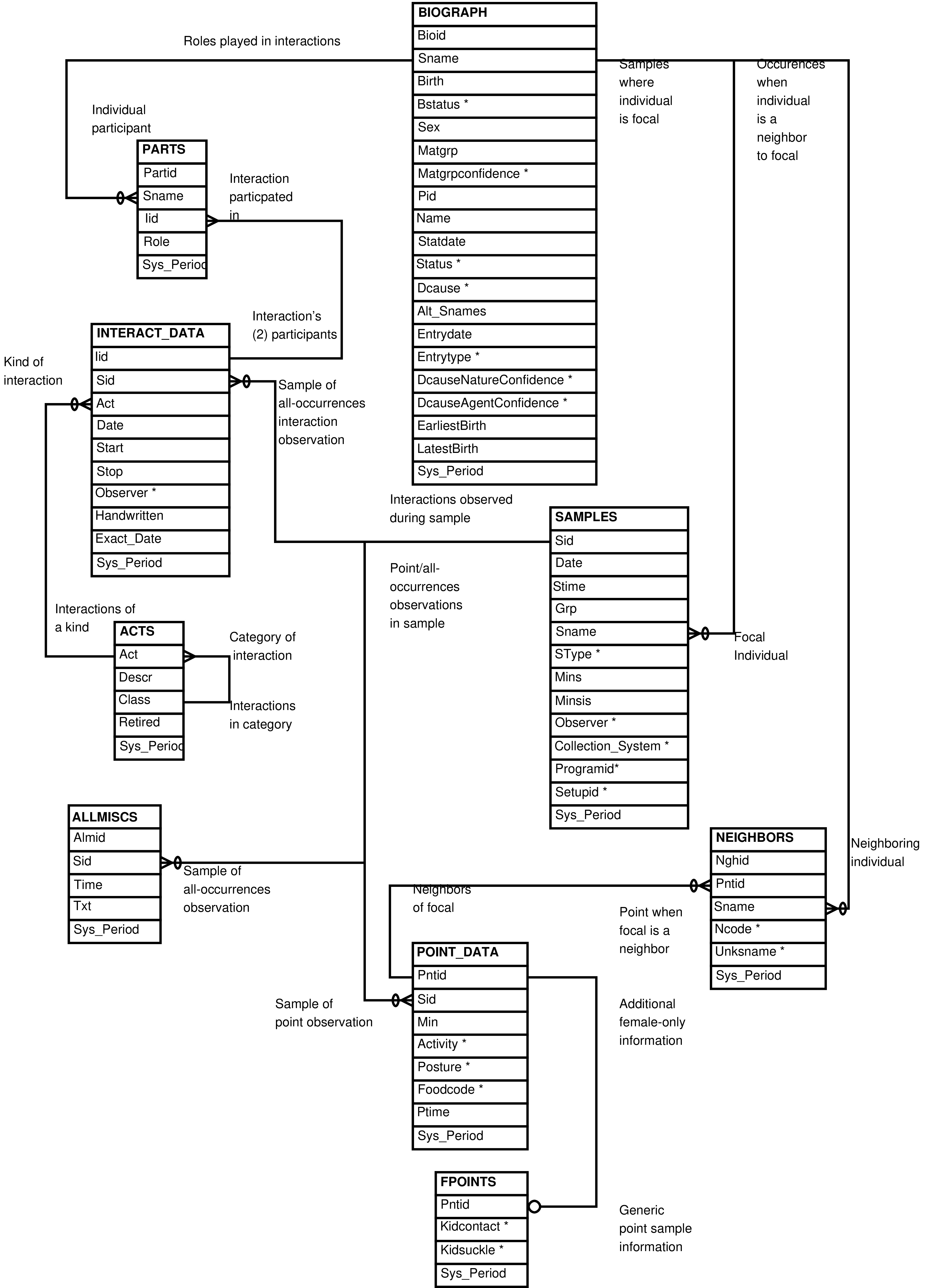
| Social and Multiparty Interactions | |
| Table | One row for each |
| ALLMISCS | “free form” all-occurrences datum |
| CONSORTS | multiparty dispute over a consortship |
| FPOINTS | point observation of a mature female |
| INTERACT_DATA | interaction between individuals |
| MPIS | collection of multiparty interactions |
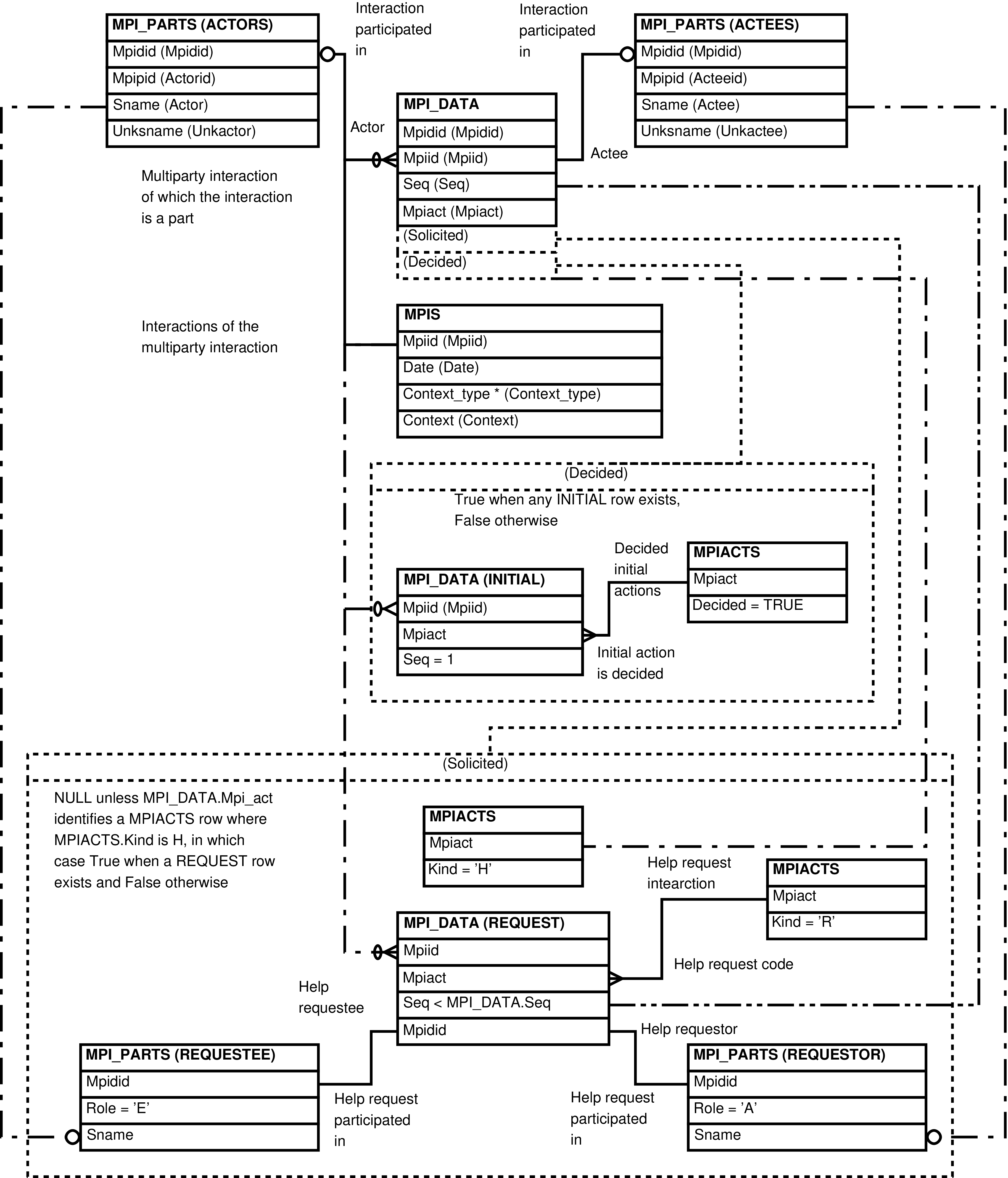
| MPI_DATA | single dyadic interaction of a multiparty interaction collection |
| MPI_PARTS | participant in a dyadic interaction of a multiparty interaction collection |
| PARTS | participant in each interaction |
| POINT_DATA | individual point observation |
| NEIGHBORS | neighbor recorded in each point sample |
| SAMPLES | focal sample |
| Darting | |
| Table | One row for each |
| ANESTHS | time additional sedation is administered to a darted individual |
| BODYTEMPS | body temperature measurement taken of a darted individual |
| CHESTS | chest circumference measurement made of a darted individual |
| CROWNRUMPS | crown to rump measurement made of a darted individual |
| DART_SAMPLES | sample type collected at each darting |
| DARTINGS | darting of an animal when data was collected |
| DPHYS | darting event during which physiological measurements were taken |
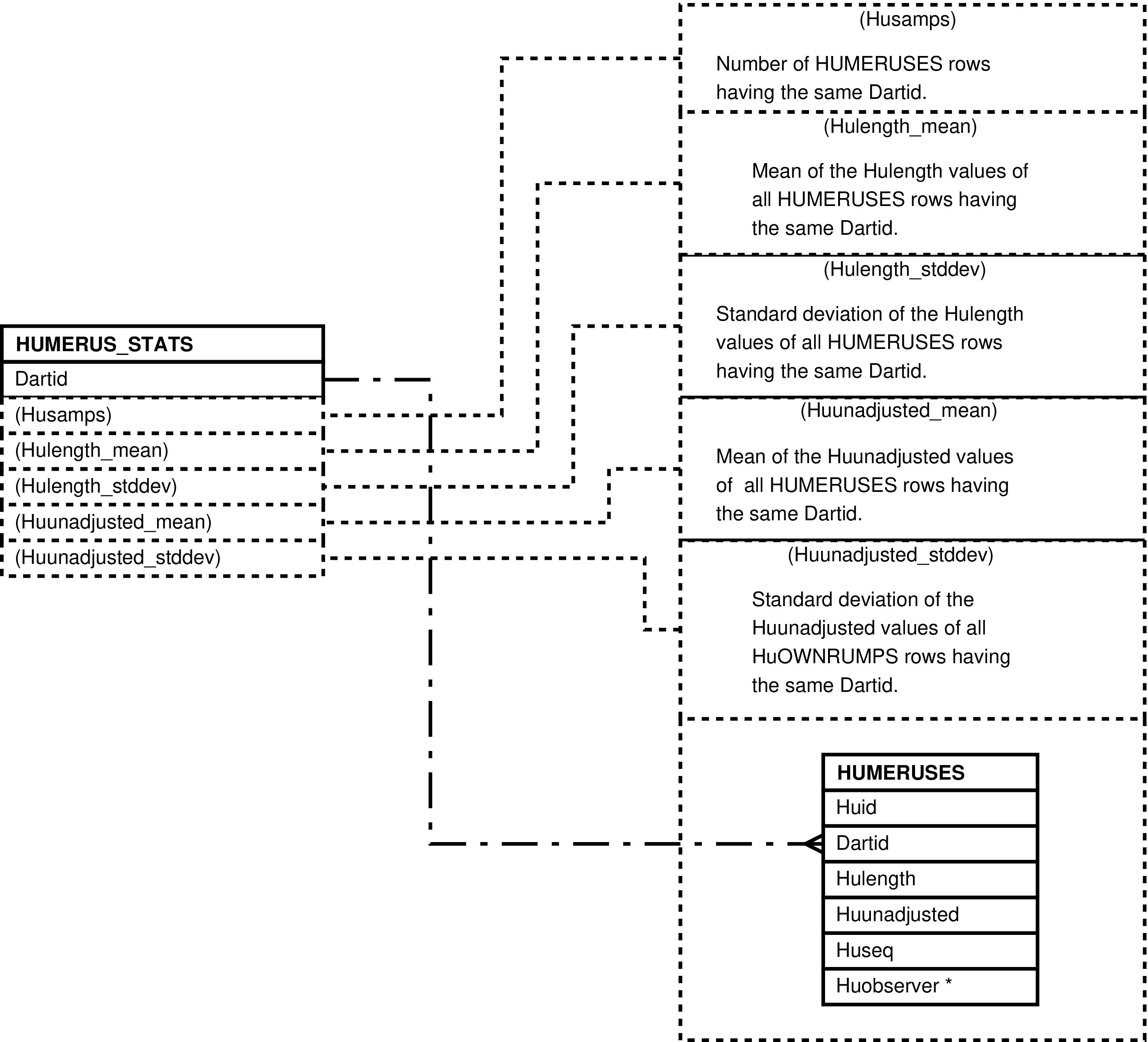
| HUMERUSES | humerous length measurement made of a darted individual |
| PCVS | packed cell volume measurement taken from a darted individual |
| TEETH | possible tooth site within the mouth on which data was collected for every darting event during which dentition data was collected |
| TESTES_ARC | every testicle width/length measurement recorded, as measured along a portion of the circumference |
| TESTES_DIAM | every testicle width/length measurement recorded, as measured along the diameter |
| TICKS | darting event during which data on ticks and other parasites were recorded |
| ULNAS | ulna length measurement made of a darted individual |
| VAGINAL_PHS | vaginal pH measurement made of a darted individual |
| Analyzed: Darting | |
| Table | One row for each |
| FLOW_CYTOMETRY | flow cytometric analysis of a blood sample collected during a darting |
| WBC_COUNTS | count from a blood smear collected during a darting |
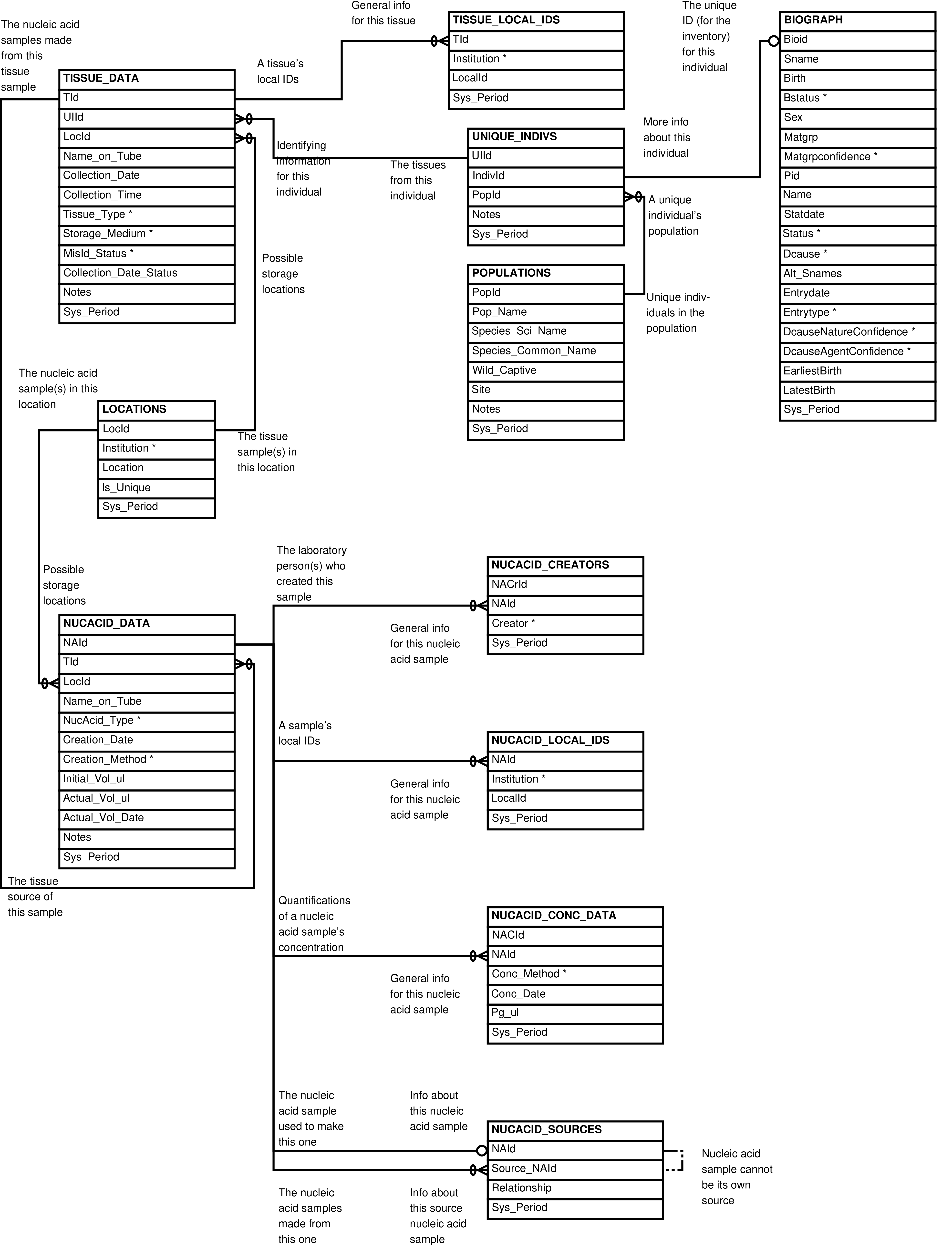
| Inventory | |
| Table | One row for each |
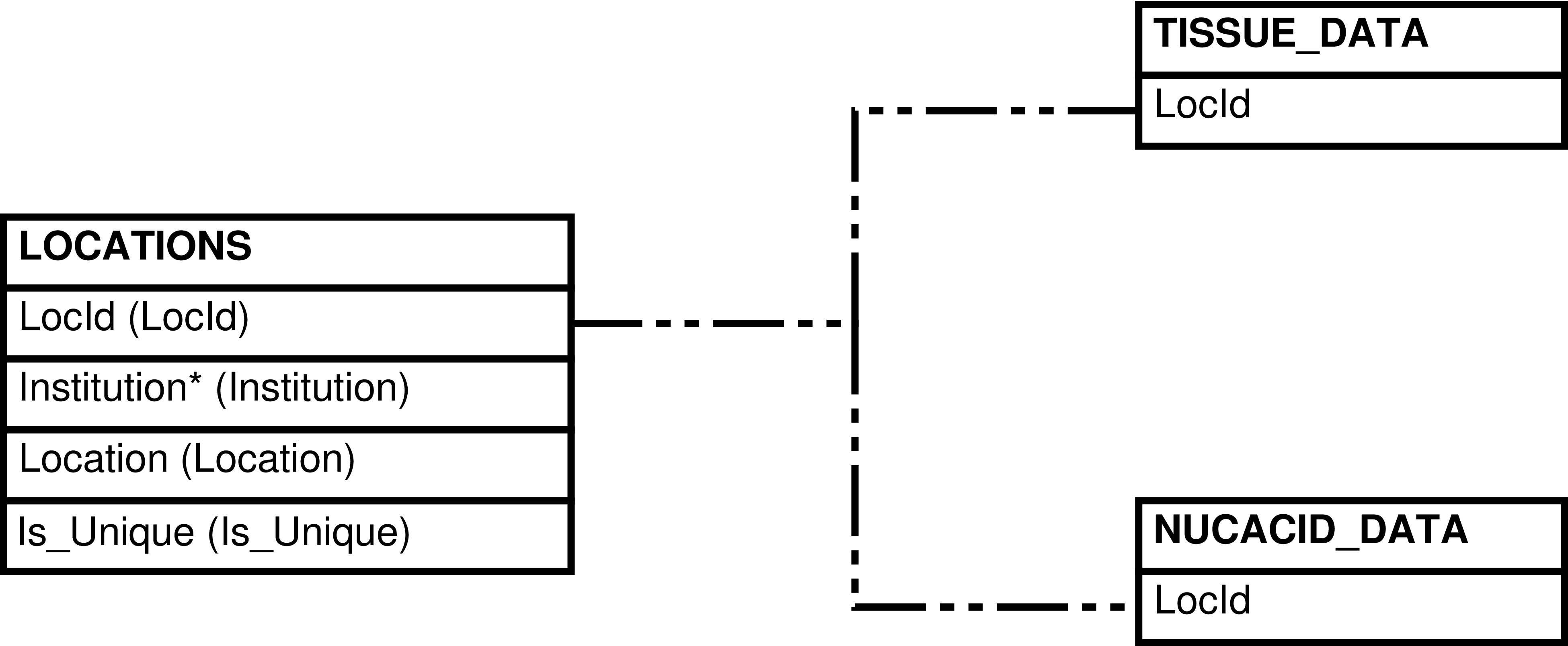
| LOCATIONS | Location that can be used to store tissue and nucleic acid samples |
| NUCACID_CONC_DATA | Quantification of a nucleic acid sample's concentration |
| NUCACID_DATA | Nucleic acid sample that is or ever has been in the inventory |
| NUCACID_LOCAL_IDS | Name/ID used to identify a nucleic acid sample at a particular institution |
| NUCACID_SOURCES | Nucleic acid sample that has another nucleic acid sample as its source |
| POPULATIONS | Study population under observation or from which tissue or nucleic acid samples have been collected |
| TISSUE_DATA | Tissue sample that is or ever has been in the inventory |
| TISSUE_LOCAL_IDS | Name/ID used to identify a tissue sample at a particular institution |
| TISSUE_SOURCES | Tissue sample that has another tissue sample as its source |
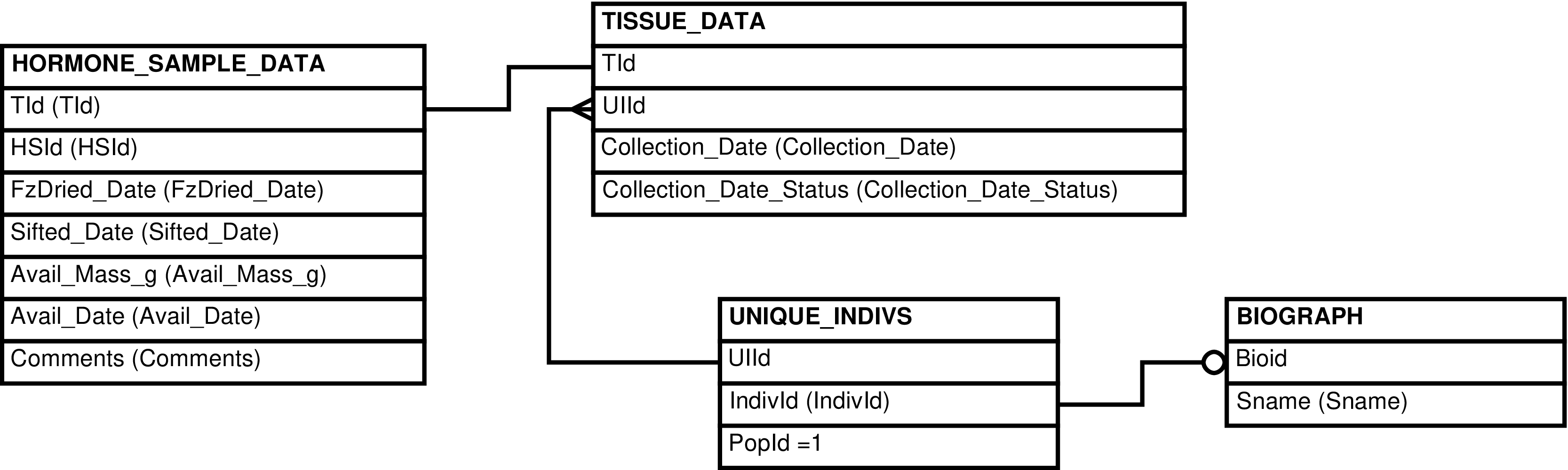
| UNIQUE_INDIVS | Individual under observation or from whom tissue or nucleic acid samples have been collected |
| SWERB Data (Group-level Geolocation Data) | |
| Table | One row for each |
| AERIALS | aerial photo used for map quadrant specification |
| GPS_UNITS | GPS device |
| QUAD_DATA | SWERB map quadrant |
| SWERB_BES | uninterrupted bout of group-level observation |
| SWERB_DATA | event related to group-level geolocation |
| SWERB_DEPARTS_DATA | departure from camp of a observation team which collected SWERB data |
| SWERB_GWS | geolocated physical object (grove or waterhole) |
| SWERB_GW_LOC_DATA | recorded location of a geolocated physical object (grove or waterhole) |
| SWERB_LOC_DATA | observation of a group at a time at a geolocated physical object |
| SWERB_LOC_DATA_CONFIDENCES | analyzed observation of a location |
| SWERB_LOC_GPS | observation of a group at a time at a geolocated physical object made using gps units and a protocol that requires 2 waypoint readings |
| SWERB_OBSERVERS | departure from camp of an observer who drove or collected SWERB data |
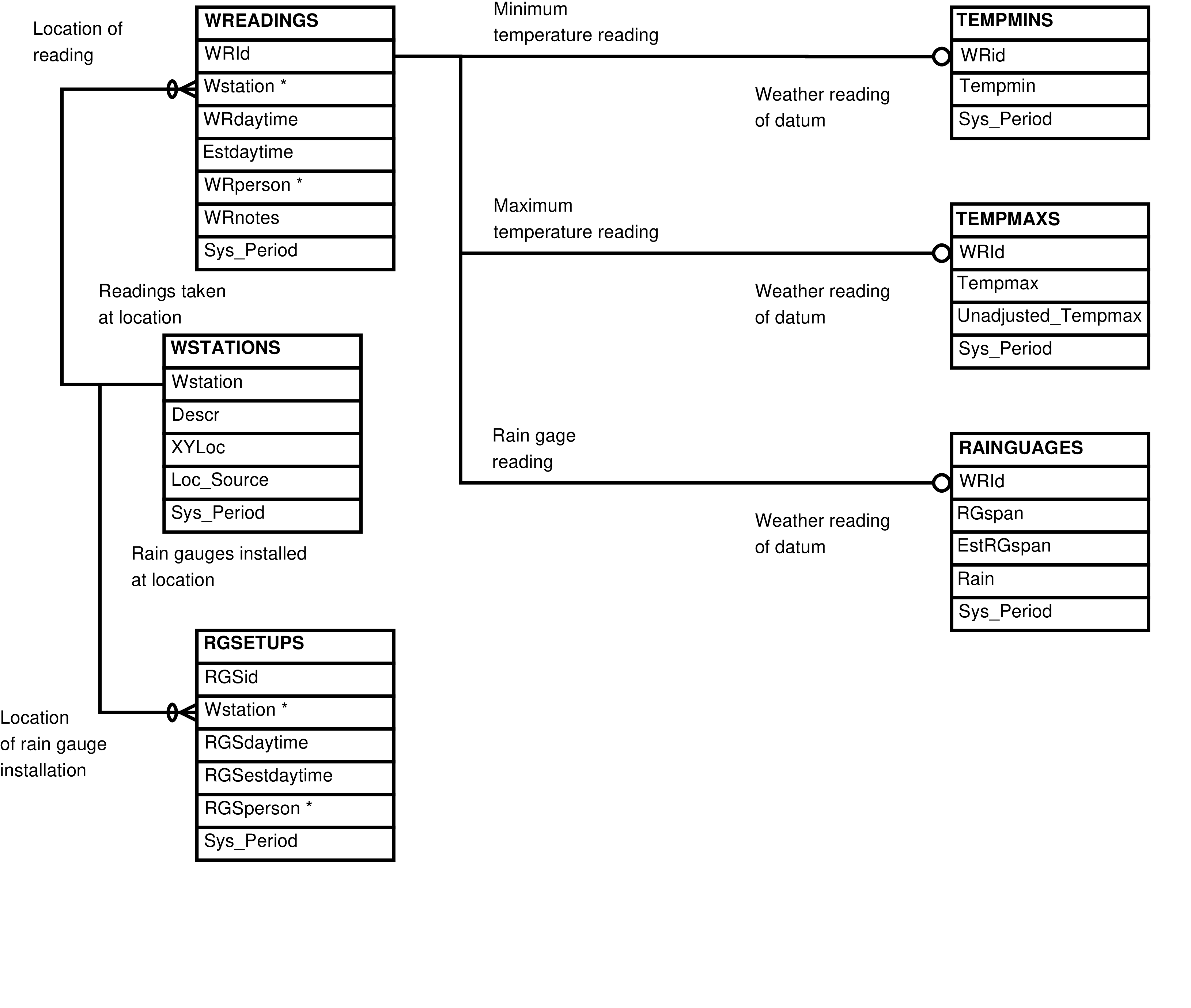
| Weather Data | |
| Table | One row for each |
| RAINGAUGES | rain gauge reading |
| RGSETUPS | rain gauge installation |
| TEMPMAXS | maximum temperature reading |
| TEMPMINS | minimum temperature reading |
| DIGITAL_WEATHER | digital weather reading reported from an electronic weather collection device |
| WREADINGS | manually collected meteorological data collection event |
| Metadata | |
| Table | One row for each |
| BEHAVE_GAPS | gap in observation |
Table 2. The Babase Support Tables
| General Support Tables | |||
| Table | Id Column | Related Column(s) | One entry for every possible choice of... |
| BODYPARTS | Bodypart | TICKS.Bodypart, BODYPARTS.Bodyregion, WP_AFFECTEDPARTS.Bodypart | part of the body |
| LAB_PERSONNEL | Initials | HYBRIDGENE_ANALYSES.Analyzed_By, NUCACID_CREATORS.Creator, WBC_COUNTS.Counted_By | person who generates data, usually in a lab setting |
| OBSERVERS | Initials | SAMPLES.Observer, WREADINGS.WRperson, RGSETUPS.RGSPerson, CROWNRUMPS.CRobserver, CHESTS.Chobserver, ULNAS.Ulobserver, HUMERUSES.Huobserver, SWERB_OBSERVERS.Observer | person who record observational data |
| OBSERVER_ROLES | Initials | OBSERVERS.Role, OBSERVERS.SWERB_Observer_Role, OBSERVERS.SWERB_Driver_Role, SWERB_OBSERVERS.Role | way in which a person can be involved in the data collection process |
| UNKSNAMES | Unksname | NEIGHBORS.Unksname and the SWERB_UPLOAD view | problem in identifying neighbor of focal during point sampling or in identifying a lone male in a SWERB other group observation |
| Group Membership and Life Events | |||
| Table | Id Column | Related Column(s) | One entry for every possible choice of... |
| BSTATUSES | Bstatus | BIOGRAPH.Bstatus | birthday estimation accuracy |
| CONFIDENCES | Confidence | BIOGRAPH.DcauseNatureConfidence, BIOGRAPH.DcauseAgentConfidence, DISPERSEDATES.Dispconfidence, BIOGRAPH.Matgrpconfidence | degree of certitude in nature of death, agent of death, disperse date assignment, or maternal group assignment |
| DADS_COMPLETENESS | Completeness | DADS_CONSENSUS.Completeness | category of analysis completeness |
| DADS_EVIDENCE_TERMS | Term | DADS_EVIDENCE.Term | kind of evidence used in a paternity analysis |
| DADS_MISMATCHES | Mismatch | DADS_CONSENSUS.Mismatch | category of analysis completeness |
| DCAUSES | Dcause | BIOGRAPH.Dcause | cause of death |
| DEATHNATURES | Nature | DCAUSES.Nature | reason for death |
| DEMOG_REFERENCES | Reference | DEMOG.Reference | data source for demography notes |
| MSTATUSES | Mstatus | MATUREDATES.Matured, RANKDATES.Ranked | maturity marker date estimation process |
| RNKTYPES | Rnktype | RANKS.Rnktype | rank ordering assigned to subject and month |
| STATUSES | Status | BIOGRAPH.Status | baboon alive at last observation |
| Physical Traits | |||
| Table | Id Column | Related Column(s) | One entry for every possible choice of... |
| HORMONE_IDS | Hormone | HORMONE_KITS.Hormone | hormone that may be extracted and assayed for |
| HORMONE_PREP_PROCEDURES | Id | HORMONE_PREP_DATA.Procedure | procedure that may be performed in preparation for a hormone assay |
| HYBRIDGENE_SOFTWARE | Software | HYBRIDGENE_ANALYSES.Software | software used for genetic hybrid score analysis |
| MARKERS | Marker | HYBRIDGENE_ANALYSES.Marker | type of genetic marker used for genetic hybrid score analysis |
| WP_HEALSTATUSES | Healstatus | WP_HEALUPDATES.HealStatus | healing progress used in healing updates |
| WP_REPORTSTATES | ReportState | WP_REPORTS.ReportState | status of wound/pathology report |
| WP_WOUNDPATHCODES | WoundPathCode | WP_DETAILS.WoundPathCode | wound or pathology |
| Social and Multiparty Interactions | |||
| Table | Id Column | Related Column(s) | One entry for every possible choice of... |
| ACTIVITIES | Activity | POINT_DATA.Activity | activity classification |
| ACTS | Act | INTERACT_DATA.Act | interaction classification |
| DATA_STRUCTURES | Data_Structure | SETUPIDS.Data_Structure | version of data structure produced by the data collection devices |
| CONTEXT_TYPES | Context_type | MPIS.Context_type | context in which a multiparty interaction occurs |
| FOODCODES | Foodcode | POINT_DATA.Foodcode | name of a food item |
| FOODTYPES | Ftype | FOODCODES.Ftype | food category |
| KIDCONTACTS | Kidcontact | FPOINTS.Kidcontact | spatial relationship between mother and infant |
| MPIACTS | Mpiact | MPI_DATA.MPIAct | multiparty interaction classification |
| NCODES | Ncode | NEIGHBORS.Ncode | neighbor classification |
| PARTUNKS | Unksname | MPI_PARTS.Unksname | problem in identifying participant in a multiparty interaction |
| POSTURES | Posture | POINT_DATA.Posture | designated posture |
| PROGRAMIDS | Programid | SAMPLES.Programid | version of each program used on the devices to collect focal sampling data |
| SAMPLES_COLLECTION_SYSTEMS | Collection_System | SAMPLES.Collection_System | device or "system" used in the field for collecting focal sampling data |
| SETUPIDS | Setupid | SAMPLES.Setupid | setupfile used on the devices to collect focal sampling data |
| STYPES | SType | SAMPLES.SType | protocol for focal sampling data collection |
| STYPES_ACTIVITIES | SType-Activity pair | SAMPLES.SType, ACTIVITIES.Activity | activity classification allowed to be used in each focal sampling protocol |
| STYPES_NCODES | SType-Ncode pair | SAMPLES.SType, NCODES.Ncode | neighbor classification allowed to be used in each focal sampling protocol |
| STYPES_POSTURES | SType-Posture pair | SAMPLES.SType, POSTURES.Posture | posture classification allowed to be used in each focal sampling protocol |
| SUCKLES | Suckle | FPOINTS.Kidsuckle | infant suckling activity |
| Sexual Cycles and The Sexual Cycle Day-By-Day Tables | |||
| Table | Id Column | Related Column(s) | One entry for every possible choice of... |
| PCSCOLORS | Color | SEXSKINS.Color | paracallosal skin coloration |
| Darting | |||
| Table | Id Column | Related Column(s) | One entry for every possible choice of... |
| DART_SAMPLE_CATS | Ds_cat | DART_SAMPLE_CATS.DS_Cat | category of darting sample type |
| DART_SAMPLE_TYPES | DS_Type | DART_SAMPLE_TYPES.DS_Type | type of sample collected during dartings |
| DRUGS | Drug | DRUGS.Drug | anesthetic drug |
| LYMPHSTATES | Lymphstate | DPHYS.Ringnode, DPHYS.Lingnode, DPHYS.Raxnode, DPHYS.Laxnode, DPHYS.Lsubmandnode, DPHYS.Rsubmandnode | lymph node condition |
| PARASITES | PARASITE | TICKS.Tickkind | parasite species, species developmental stage, or kind of parasite sign counted |
| TCONDITIONS | Tcondition | TEETH.Tcondition | physical condition of a tooth |
| TICKSTATUSES | Tickstatus | TICKS.Tickstatus | parasite count outcome category |
| TOOTHCODES | Tooth | TEETH.Tooth | adult or deciduous tooth |
| TOOTHSITES | Toothsite | TOOTHCODES.Toothsite | dental site within the mouth |
| TSTATES | Tstate | TEETH.Tstate | tooth “presence” |
| Inventory | |||
| Table | Id Column | Related Column(s) | One entry for every possible ... |
| INSTITUTIONS | Institution | LOCATIONS.Institution, NUCACID_LOCAL_IDS.Institution, TISSUE_LOCAL_IDS.Institution | possible locale where tissue and nucleic acid samples can be stored or used |
| MISID_STATUSES | Misid_Status | TISSUE_DATA.Misid_Status | level of confidence in the identity of a tissue sample |
| NUCACID_CONC_METHODS | Conc_Method | NUCACID_CONC_DATA.Conc_Method | method used for quantifying nucleic acid concentrations |
| NUCACID_CREATION_METHODS | Creation_Method | NUCACID_DATA.Creation_Method | method used for creating nucleic acid samples |
| NUCACID_TYPES | NucAcid_Type | NUCACID_DATA.NucAcid_Type | type of nucleic acid sample |
| STORAGE_MEDIA | Storage_Medium | TISSUE_DATA.Storage_Medium | medium used for storage/archiving of tissue samples |
| TISSUE_TYPES | Tissue_Type | TISSUE_DATA.Tissue_Type | type of tissue sample |
| SWERB Data (Group-level Geolocation Data) | |||
| Table | Id Column | Related Column(s) | One entry for every possible ... |
| ADCODES | ADCode | SWERB_LOC_DATA.ADcode | relationship between baboon groups and sleeping groves. |
| SWERB_LOC_CONFIDENCES | Conf | SWERB_LOC_DATA_CONFIDENCES.Confidence | confidence score used when analyzing the accuracy of a recorded observation of a location. |
| SWERB_LOC_STATUSES | Conf | SWERB_LOC_DATA.Loc_Status | status for a recorded observation of a location. |
| SWERB_TIME_SOURCES | Source | SWERB_BES.Bsource, SWERB_BES.Esource | data source used to estimate beginning and ending of observation bouts |
| SWERB_XYSOURCES (SWERB Time Sources) | Source | SWERB_GW_LOC_DATA.XYSource | data source used to obtain XY coordinates |
| Weather Data | |||
| Table | Id Column | Related Column(s) | One entry for every possible choice of... |
| WEATHER_SOFTWARES | WSoftware | DIGITAL_WEATHER.WSoftware | software used to retrieve data from an electronic weather collection instrument |
| WSTATIONS | Wstation | WREADINGS.Wstation | meteorological data collection location or device |
| Metadata | |||
| Table | Id Column | Related Column(s) | One entry for every possible choice of... |
| GAP_END_STATUSES | Gap_End_Status | BEHAVE_GAPS.Gap_End_Status | reason why an observation gap ended |
Table 3. The Babase Views
| Group Membership and Life Events | |||
| View | One row for each | Purpose | Tables/Views used |
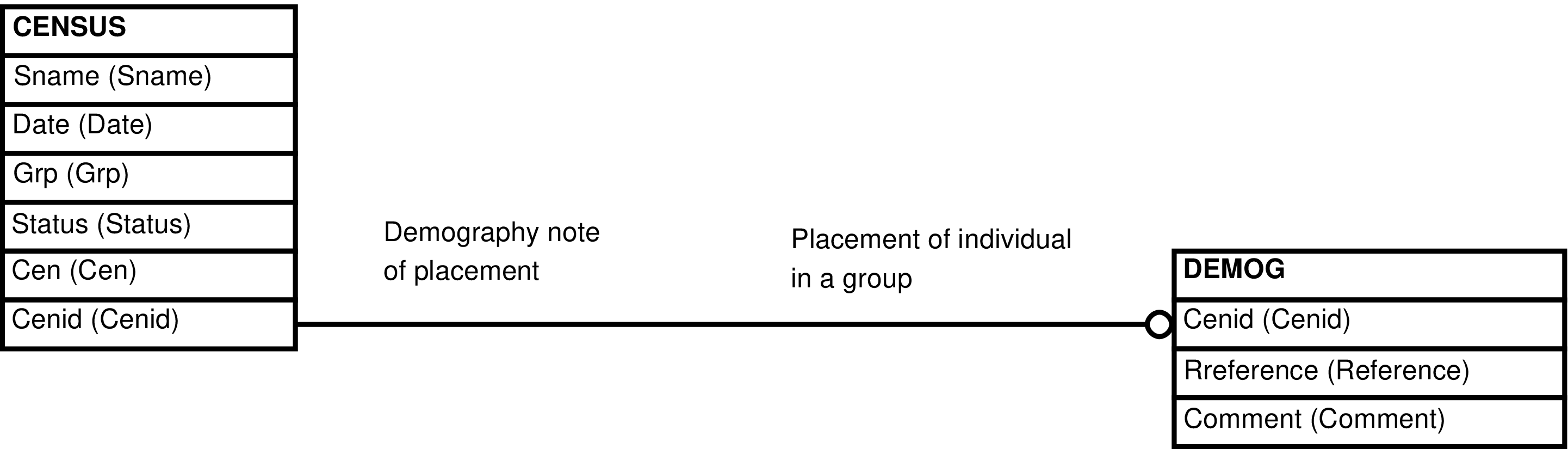
| CENSUS_DEMOG | CENSUS row | Maintenance of CENSUS rows that are extended with DEMOG information. | CENSUS, DEMOG |
| CENSUS_DEMOG_SORTED | CENSUS row | Maintenance of CENSUS_DEMOG rows in a pre-sorted fashion. | CENSUS, DEMOG |
| CYCPOINTS_CYCLES | CYCPOINTS row | Maintenance of CYCPOINTS rows that are extended with CYCLES information. | CYCLES, CYCPOINTS |
| CYCPOINTS_CYCLES_SORTED | CYCPOINTS row | The CYCPOINTS_CYCLES view sorted by CYCLES.Sname, by CYCPOINTS.Date. | CYCLES, CYCPOINTS |
| DEMOG_CENSUS | DEMOG row | Maintenance of DEMOG rows. | CENSUS, DEMOG |
| DEMOG_CENSUS_SORTED | CENSUS row | Maintenance of DEMOG_CENSUS rows in a pre-sorted fashion. | CENSUS, DEMOG |
| GROUPS_HISTORY | GROUPS row | Depiction of GROUPS rows in a more human-readable format. | GROUPS |
| PARENTS | BIOGRAPH
row for which there is either a row in MATERNITIES with a
record of the individual's mother or there is a row in DADS_CONSENSUS with a
record of the individual's father -- with a non-NULLDad_Consensus. | Easy access to parental information. | BIOGRAPH, MATERNITIES, DADS_CONSENSUS, MEMBERS |
| POTENTIAL_DADS | (completed) female reproductive event for every male more than 2192 days old (approximately 6 years) present in the mother's group during her fertile period | Research into paternity, especially the selection of potential fathers for further genetic testing. | MATERNITIES, MEMBERS (multiple times), ACTOR_ACTEES (multiple times), BIOGRAPH, RANKDATES, MATUREDATES |
| PROPORTIONAL_RANKS | RANKS row | Automatic calculation of proportional ranks from the ordinal ranks in RANKS. | RANKS |
| Physical Traits | |||
| View | One row for each | Purpose | Tables/Views used |
| ESTROGENS | HORMONE_RESULT_DATA row with an estrogen kit | Easy access to estrogen data. | BIOGRAPH, HORMONE_KITS, HORMONE_PREP_DATA, HORMONE_PREP_SERIES, HORMONE_RESULT_DATA, HORMONE_SAMPLE_DATA, TISSUE_DATA, UNIQUE_INDIVS |
| GLUCOCORTICOIDS | HORMONE_RESULT_DATA row with a glucocorticoid kit. | Easy access to glucocorticoid data. | BIOGRAPH, HORMONE_KITS, HORMONE_PREP_DATA, HORMONE_PREP_SERIES, HORMONE_RESULT_DATA, HORMONE_SAMPLE_DATA, TISSUE_DATA, UNIQUE_INDIVS |
| HORMONE_PREPS | HORMONE_PREP_DATA row | Presents HORMONE_PREP_DATA with identifying information from TISSUE_DATA and BIOGRAPH. Also useful for maintaining data in HORMONE_PREP_DATA. | BIOGRAPH, HORMONE_PREP_DATA, HORMONE_PREP_SERIES, HORMONE_SAMPLE_DATA, TISSUE_DATA, UNIQUE_INDIVS |
| HORMONE_RESULTS | HORMONE_RESULT_DATA row | Presents HORMONE_RESULT_DATA with identifying information from TISSUE_DATA and BIOGRAPH. Also useful for maintaining data in HORMONE_RESULT_DATA. | BIOGRAPH, HORMONE_KITS, HORMONE_PREP_SERIES, HORMONE_RESULT_DATA, HORMONE_SAMPLE_DATA, TISSUE_DATA, UNIQUE_INDIVS |
| HORMONE_SAMPLES | HORMONE_SAMPLE_DATA row | Presents HORMONE_SAMPLE_DATA with identifying information from TISSUE_DATA and BIOGRAPH. Also useful for maintaining data in HORMONE_SAMPLE_DATA. | BIOGRAPH, HORMONE_SAMPLE_DATA, TISSUE_DATA, UNIQUE_INDIVS |
| HYBRIDMORPH_SCORES | HYBRIDMORPH_REPORTS row | Viewing and uploading morphological hybrid score data. | HYBRIDMORPH_OBSERVERS, HYBRIDMORPH_REPORTS, HYBRIDMORPH_SCORE_DATA |
| PROGESTERONES | HORMONE_RESULT_DATA row with a progesterone kit. | Easy access to progesterone data. | BIOGRAPH, HORMONE_KITS, HORMONE_PREP_DATA, HORMONE_PREP_SERIES, HORMONE_RESULT_DATA, HORMONE_SAMPLE_DATA, TISSUE_DATA, UNIQUE_INDIVS |
| TESTOSTERONES | HORMONE_RESULT_DATA row with a testosterone kit. | Easy access to testosterone data. | BIOGRAPH, HORMONE_KITS, HORMONE_PREP_DATA, HORMONE_PREP_SERIES, HORMONE_RESULT_DATA, HORMONE_SAMPLE_DATA, TISSUE_DATA, UNIQUE_INDIVS |
| THYROID_HORMONES | HORMONE_RESULT_DATA row with a thyroid hormone kit. | Easy access to thyroid hormone data. | BIOGRAPH, HORMONE_KITS, HORMONE_PREP_DATA, HORMONE_PREP_SERIES, HORMONE_RESULT_DATA, HORMONE_SAMPLE_DATA, TISSUE_DATA, UNIQUE_INDIVS |
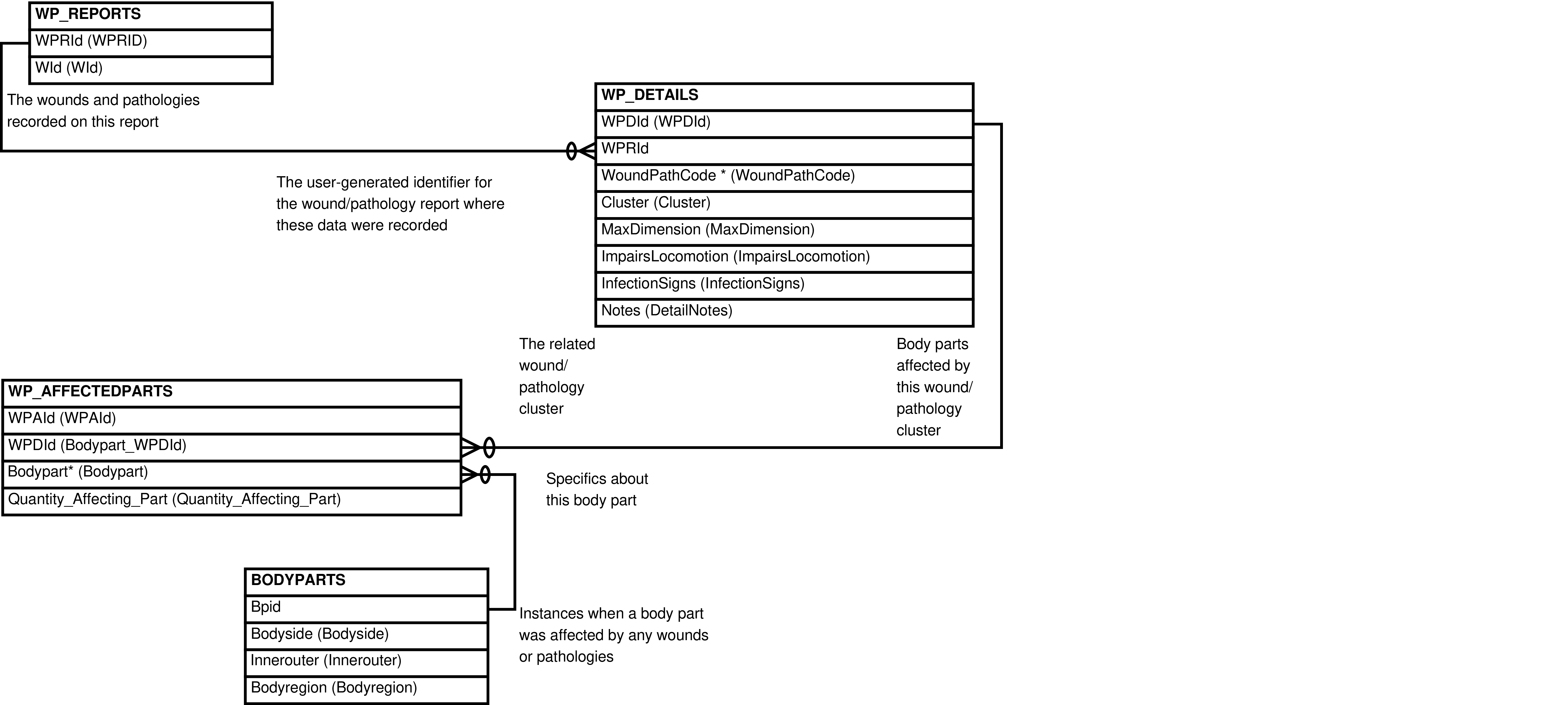
| WOUNDSPATHOLOGIES | WP_AFFECTEDPARTS row | Querying of wounds/pathologies data (without heal updates). | WP_REPORTS, WP_OBSERVERS, WP_DETAILS, WP_AFFECTEDPARTS, BODYPARTS |
| WP_DETAILS_AFFECTEDPARTS | WP_AFFECTEDPARTS row | Upload of WP_DETAILS and WP_AFFECTEDPARTS rows. | WP_DETAILS, WP_AFFECTEDPARTS, BODYPARTS |
| WP_HEALS | WP_HEALUPDATES row | Upload and viewing of WP_HEALUPDATES rows. | WP_REPORTS, WP_DETAILS, WP_AFFECTEDPARTS, BODYPARTS, WP_HEALUPDATES |
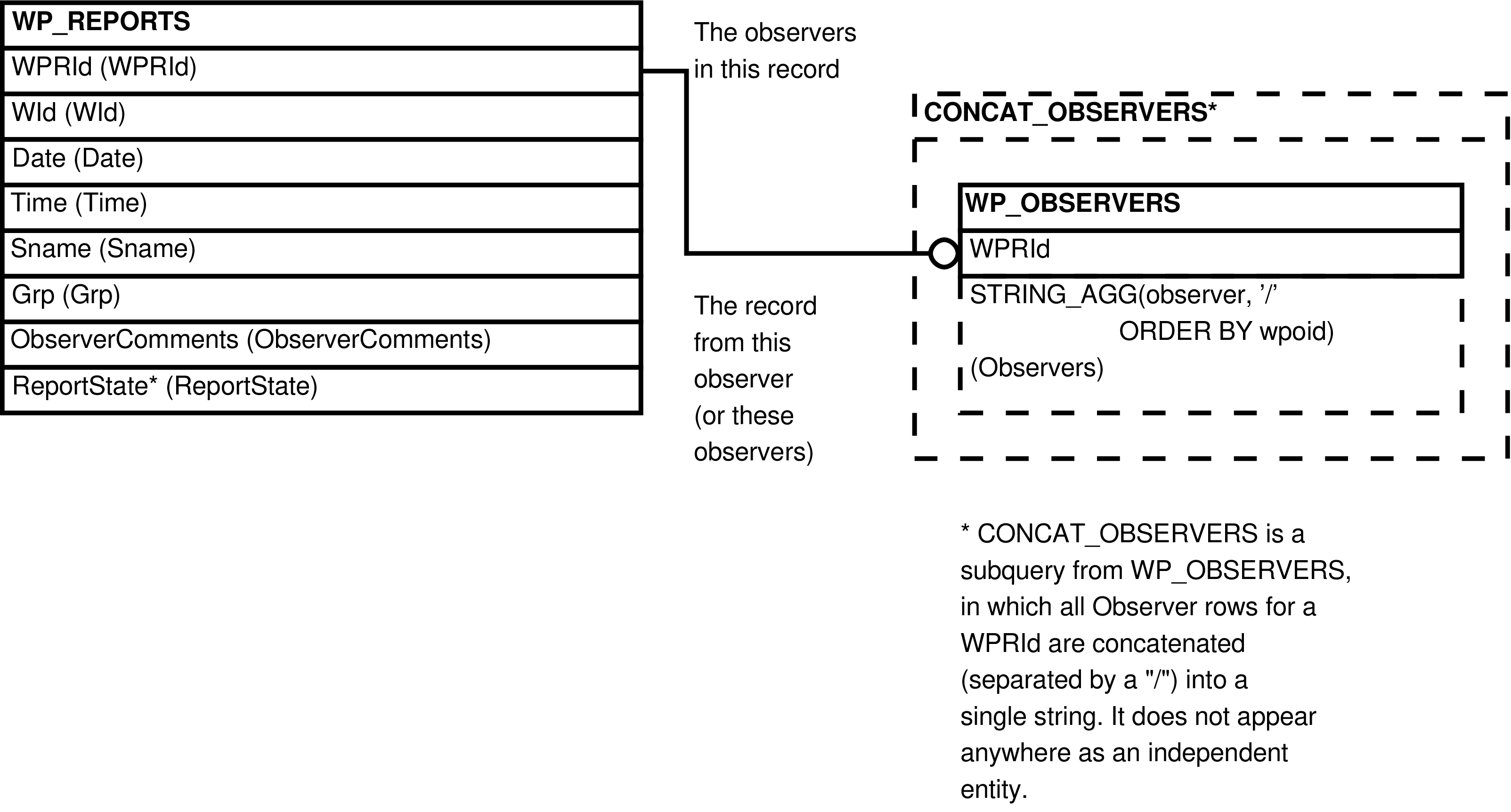
| WP_REPORTS_OBSERVERS | WP_REPORTS row | Upload of WP_REPORTS and WP_OBSERVERS rows. | WP_REPORTS, WP_OBSERVERS |
| Sexual Cycles | |||
| View | One row for each | Purpose | Tables/Views used |
| CYCLES_SEXSKINS | CYCLES row | Maintenance of SEXSKINS rows. | CYCLES, SEXSKINS |
| CYCLES_SEXSKINS_SORTED | CYCLES row | The CYCLES_SEXSKINS view sorted by CYCLES.Sname, by SEXSKINS.Date. | CYCLES, SEXSKINS |
| MATERNITIES | birth or fetal loss | Summarizes (completed) reproductive events. | BIOGRAPH, PREGS, CYCPOINTS, CYCLES |
| MTD_CYCLES | CYCLES row | Presents CYCLES together with Mdate, Tdate, and Ddate CYCPOINTS information for a view of an "entire" sexual cycle as a single row. | CYCLES, CYCPOINTS |
| SEXSKINS_CYCLES | SEXSKINS row | Maintenance of SEXSKINS rows. | CYCLES, SEXSKINS |
| SEXSKINS_CYCLES_SORTED | SEXSKINS row | The SEXSKINS_CYCLES view sorted by CYCLES.Sname, by SEXSKINS.Date. | CYCLES, SEXSKINS |
| SEXSKINS_REPRO_NOTES | SEXSKINS row, or REPRO_NOTES row | Maintenance of SEXSKINS rows. | CYCLES, REPRO_NOTES, SEXSKINS |
| Social and Multiparty Interactions | |||
| View | One row for each | Purpose | Tables/Views used |
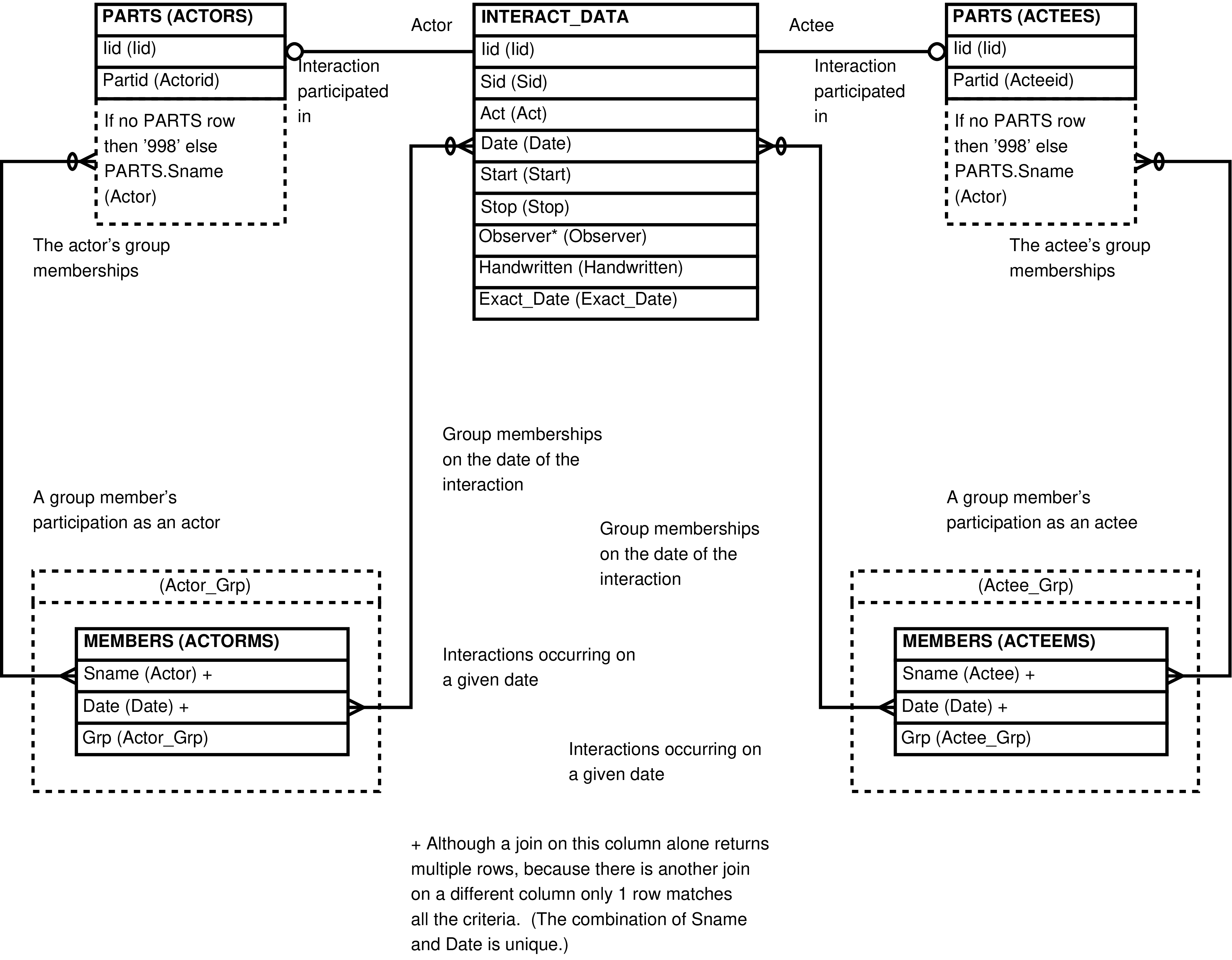
| ACTOR_ACTEES | INTERACT row | Maintenance of social interaction data, INTERACT rows and POINTS. A view optimized for highest performance when working with these tables. Analysis of social interaction data. | INTERACT, PARTS |
| INTERACT | INTERACT_DATA row | Presents INTERACT_DATA with additional date and time columns that transform the underlying date and time columns in useful and interesting ways. | INTERACT_DATA |
| INTERACT_SORTED | INTERACT_DATA row | Presents the INTERACT view sorted in a fashion expected to ease maintenance. | INTERACT_DATA |
| MPI_EVENTS | MPI_DATA row | Analysis and correction of multiparty interaction data. | MPI_DATA, MPI_PARTS, MPIACTS |
| POINTS | POINT_DATA row | Presents POINT_DATA with the Ptime column transformed into a column that may be useful and interesting. | POINT_DATA |
| POINTS_SORTED | POINTS row | Presents POINTS sorted by Sid, and within that by Ptime. | POINTS |
| SAMPLES_GOFF | SAMPLES row | Presents SAMPLES with an additional column Grp_of_focal, which has the group of the focal at the time of sampling. | SAMPLES |
| Darting | |||
| View | One row for each | Purpose | Tables/Views used |
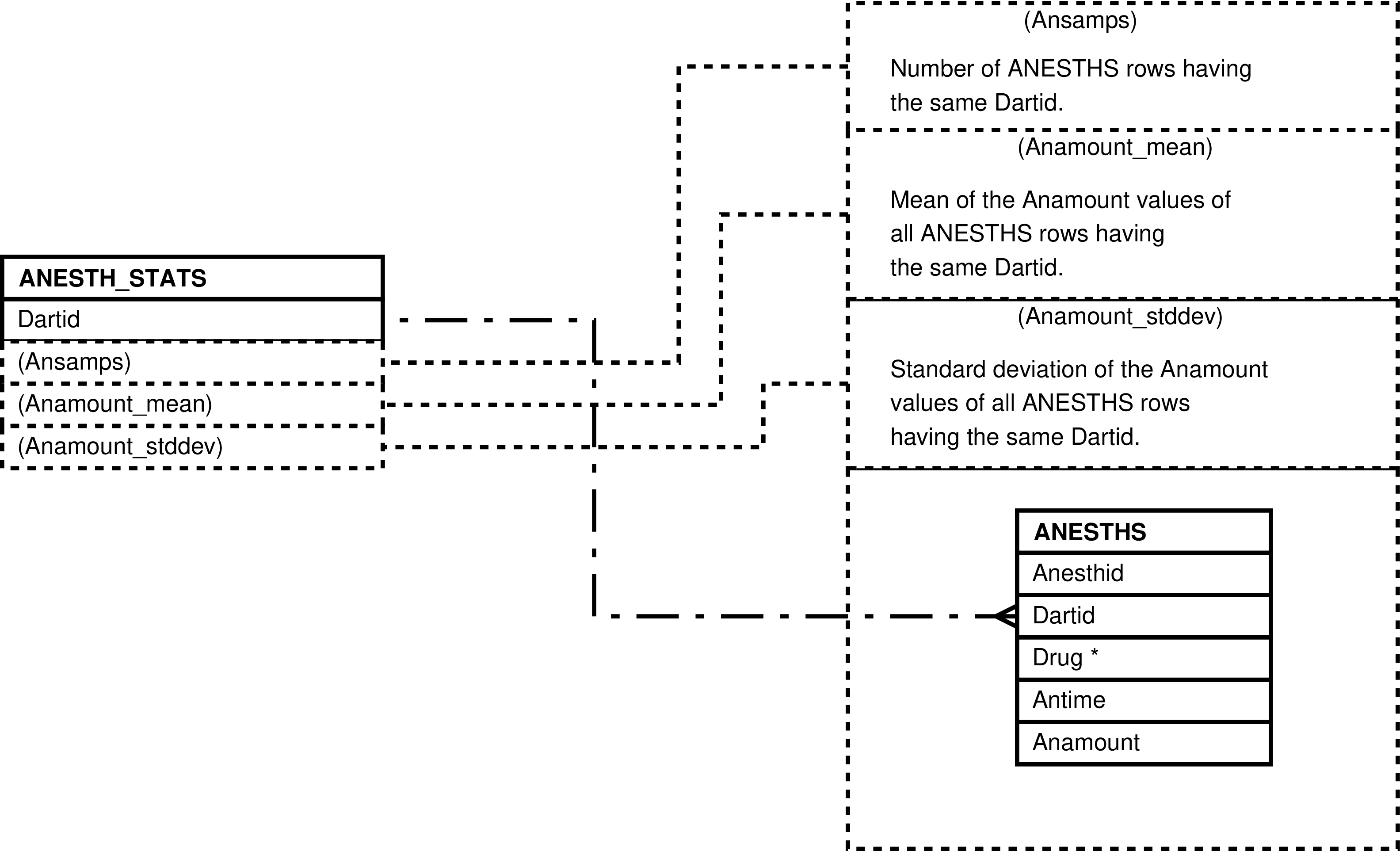
| ANESTH_STATS | unique ANESTHS.Dartid value -- for each darting during which additional anesthetic was administered | Analysis and “eyeballing” of data involving additional administration of anesthetic when darting. | ANESTHS |
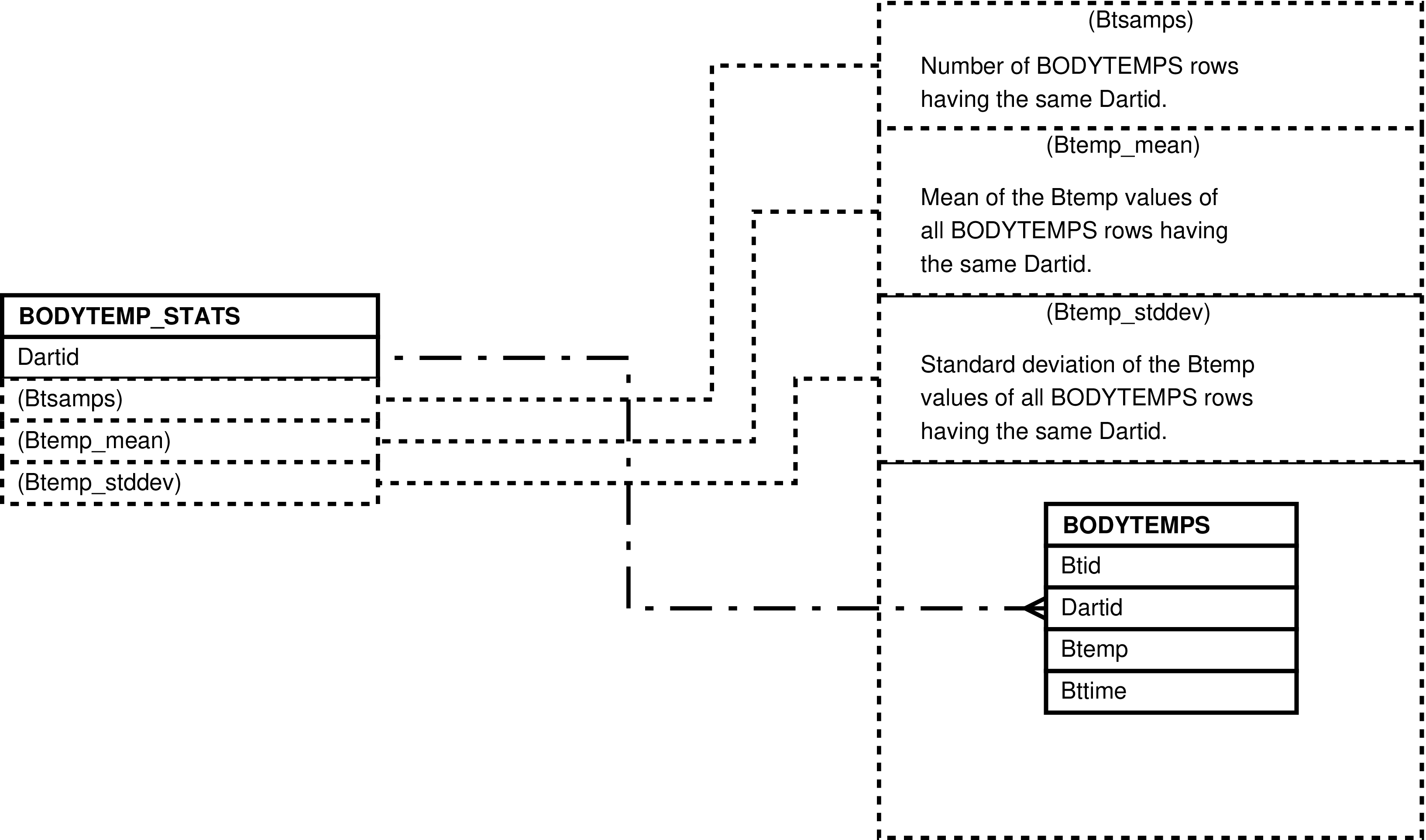
| BODYTEMP_STATS | unique BODYTEMPS.Dartid value -- for each darting having body temperature measurements | Analysis and “eyeballing” of darting body temperature measurements. | BODYTEMPS |
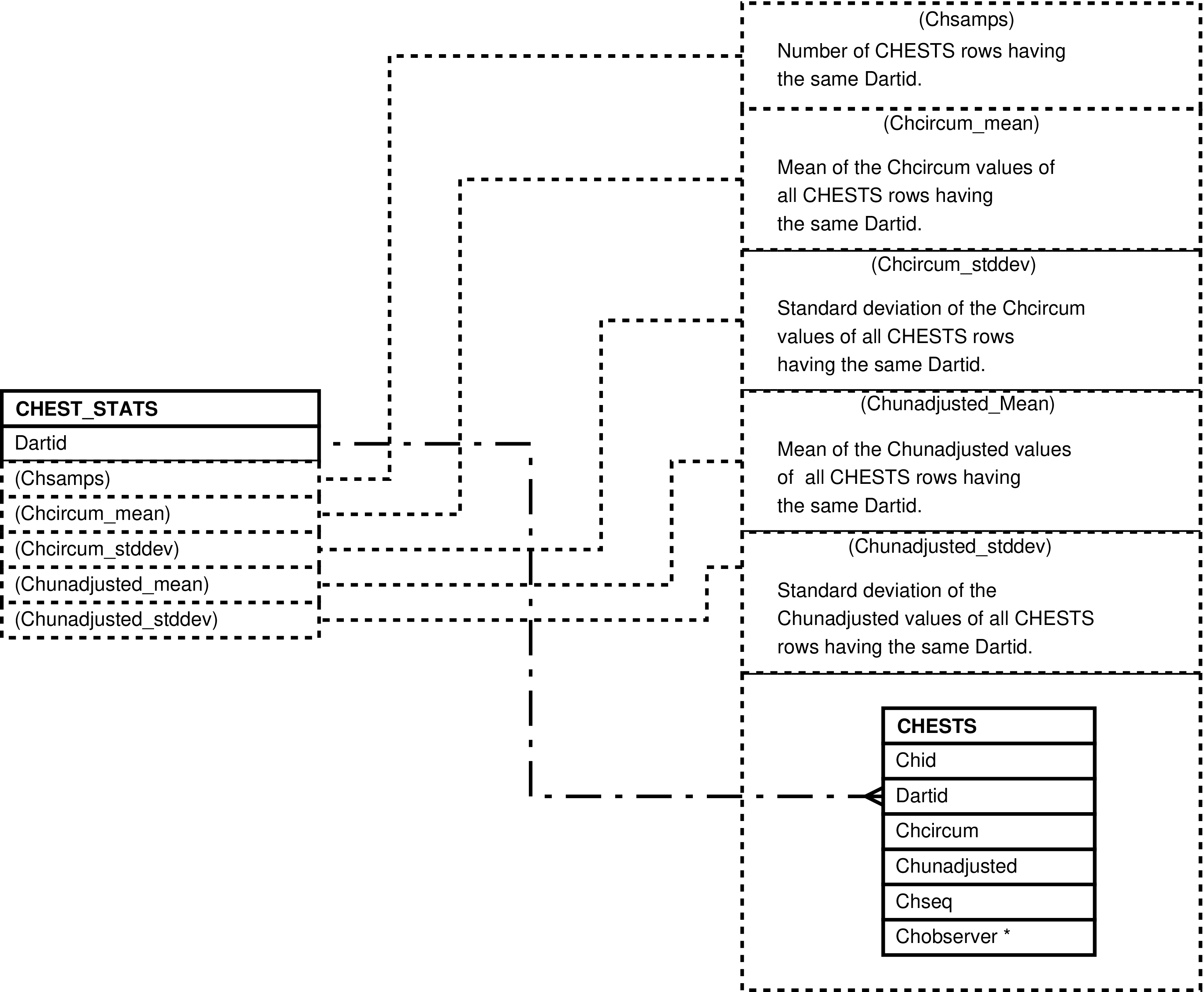
| CHEST_STATS | unique CHESTS.Dartid value -- for each darting having chest circumference measurements | Analysis and “eyeballing” of darting chest circumference measurements. | CHESTS |
| CROWNRUMP_STATS | unique CROWNRUMPS.Dartid value -- for each darting having crown-to-rump measurements | Analysis and “eyeballing” of darting crown-to-rump measurements. | CROWNRUMPS |
| DSAMPLES | unique DARTINGS.Dartid value -- for each darting | Visualization of all samples collected per darting. | DARTINGS, MEMBERS, DART_SAMPLES |
| DENT_CODES | unique TEETH.Dartid value -- for each darting with recorded tooth information | Perusal and maintenance of TEETH rows by kind of tooth. | TEETH |
| DENT_SITES | unique TEETH.Dartid value -- for each darting with recorded tooth information | Perusal of TEETH rows by position in the mouth. | TEETH, TOOTHCODES |
| HUMERUS_STATS | unique HUMERUSES.Dartid value -- for each darting having humerus length measurements | Analysis and “eyeballing” of darting humerus length measurements. | HUMERUSES |
| PCV_STATS | unique PCVS.Dartid value -- for each darting having PCV measurements | Analysis and “eyeballing” of darting PCV measurements. | PCVS |
| TESTES_ARC_STATS | unique TESTES_ARC.Dartid value -- for each darting having at least one measurement of testes length or width circumference | Analysis of testes length and width measurements taken during darting. | TESTES_ARC |
| TESTES_DIAM_STATS | unique TESTES_DIAM.Dartid value -- for each darting having at least one measurement of testes length or width diameter | Analysis of testes length and width measurements taken during darting. | TESTES_DIAM |
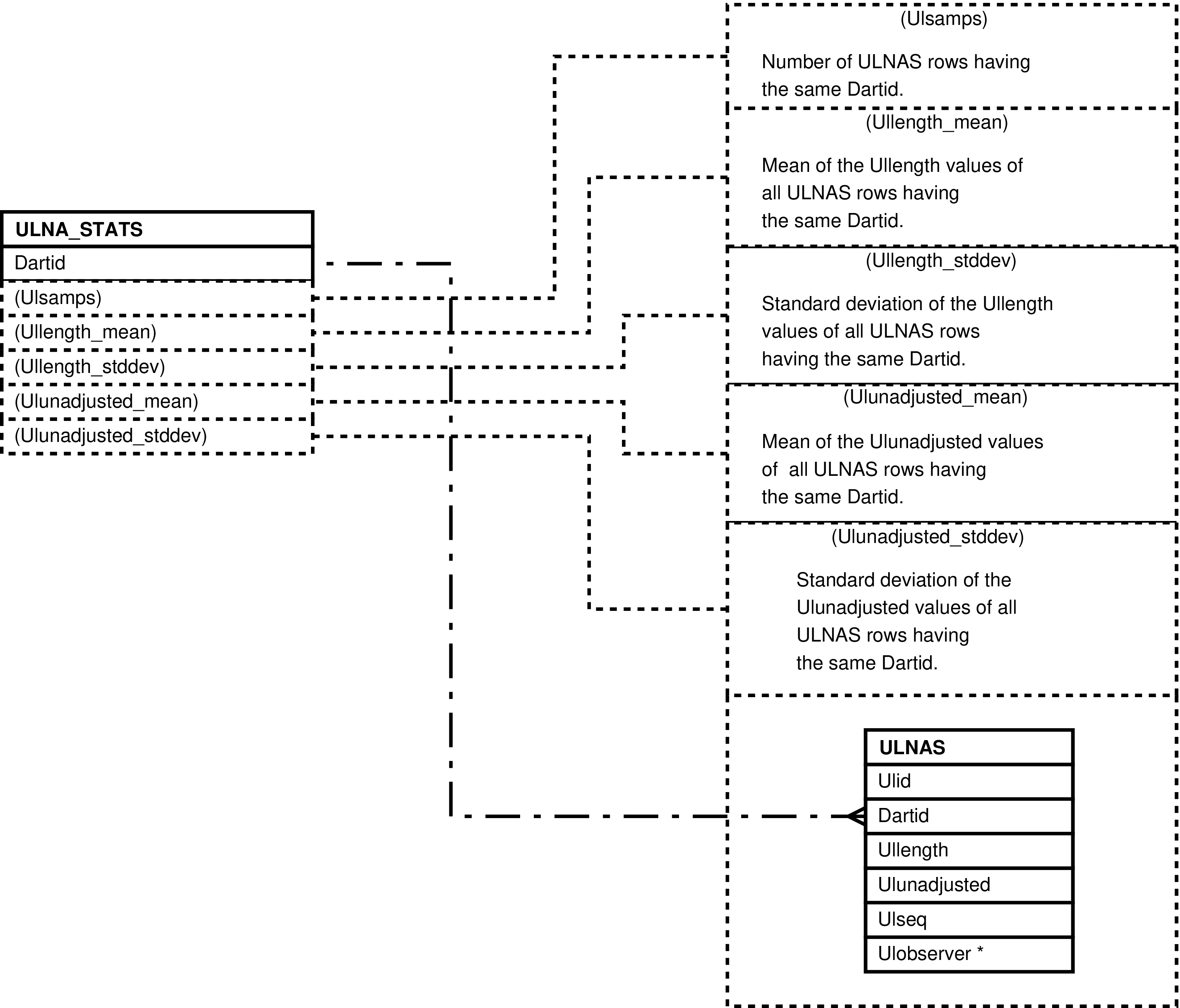
| ULNA_STATS | unique ULNAS.Dartid value -- for each darting having ulna length measurements | Analysis and “eyeballing” of darting ulna length measurements. | ULNAS |
| VAGINAL_PH_STATS | unique VAGINAL_PHS.Dartid value -- for each darting having vaginal pH measurements | Analysis and “eyeballing” of darting vaginal pH measurements. | VAGINAL_PHS |
| Inventory | |||
| View | One row for each | Purpose | Tables/Views used |
| LOCATIONS_FREE | LOCATIONS row that isn't used in NUCACID_DATA or in TISSUE_DATA | Querying of available ("free") locations for storing new samples | LOCATIONS, NUCACID_DATA, TISSUE_DATA |
| NUCACID_CONCS | NUCACID_CONC_DATA row | Converting and standardizing units of nucleic acid concentration | NUCACID_CONC_DATA, NUCACID_CONC_METHODS, NUCACID_LOCAL_IDS |
| NUCACID_SOURCES_EXT | NUCACID_SOURCES row | Viewing, inserting, and updating data about nucleic acid sources with their LocalId's | NUCACID_SOURCES, NUCACID_LOCAL_IDS |
| NUCACIDS | NUCACID_DATA row | Showing data about nucleic acids in a human-readable format | NUCACID_DATA, TISSUE_DATA, UNIQUE_INDIVS, BIOGRAPH, NUCACID_LOCAL_IDS, NUCACID_SOURCES |
| NUCACIDS_W_CONC | NUCACID_DATA row | Showing data about nucleic acids in a human-readable format, including concentrations from the most-recent quantifications | NUCACID_DATA, NUCACID_CONC_DATA, TISSUE_DATA, UNIQUE_INDIVS, BIOGRAPH, NUCACID_LOCAL_IDS, NUCACID_SOURCES |
| TISSUE_SOURCES_EXT | TISSUE_SOURCES row | Viewing, inserting, and updating data about tissue sources with their LocalId's | TISSUE_SOURCES, TISSUE_LOCAL_IDS |
| TISSUES | TISSUE_DATA row | Showing data about tissue samples in a human-readable format | TISSUE_DATA, UNIQUE_INDIVS, BIOGRAPH, TISSUE_LOCAL_IDS |
| TISSUES_HORMONES | TISSUE_DATA row | Providing an expanded set of information about tissue samples used for hormone analysis. Also useful for simultaneous upload of data to TISSUE_DATA and HORMONE_SAMPLE_DATA | TISSUE_DATA, UNIQUE_INDIVS, BIOGRAPH, TISSUE_LOCAL_IDS, HORMONE_SAMPLE_DATA |
| SWERB Data (Group-level Geolocation Data) | |||
| View | One row for each | Purpose | Tables/Views used |
| QUADS | QUAD_DATA row | Querying of X, Y coodinates from and maintenance of QUAD_DATA rows. | QUAD_DATA |
| SWERB | SWERB_DATA row -- for every SWERB event, departure from camp excluded | Collects SWERB related information spread among several tables and separates geolocation points into X and Y coordinates. | SWERB_DATA, QUADS, SWERB_BES, SWERB_DEPARTS_DATA, SWERB_DEPARTS_GPS |
| SWERB_DATA_XY | SWERB_DATA row -- for every SWERB event, departure from camp excluded | Separates SWERB_DATA geolocation points into X and Y coordinates for ease of maintenance. | SWERB_DATA |
| SWERB_DEPARTS | SWERB_DEPARTS_DATArow -- for every departure from camp of every observation team, for those observation teams which have collected SWERB data | Collects departure related information spread among several tables and separates geolocation points into X and Y coordinates. | SWERB_DEPARTS_DATA, SWERB_DEPARTS_GPS |
| SWERB_GW_LOCS | SWERB_GW_LOC_DATA row -- for every geolocation of an object, of a grove or waterhole | Collects SWERB grove and waterhole location information spread between tables and separates geolocation points into X and Y coordinates. | SWERB_GW_LOC_DATA, QUADS |
| SWERB_GW_LOC_DATA_XY | SWERB_GW_LOC_DATA row -- for every geolocation of an object, of a grove or waterhole | Separates SWERB_GW_LOC_DATA geolocation points into X and Y coordinates for ease of maintenance. | SWERB_GW_LOC_DATA |
| SWERB_LOC_GPS_XY | SWERB_LOC_GPS row -- for every time a group is observed at a geolocated physical object, usually a grove or waterhole, and 2 GPS waypoints are required to by the protocol to collect the data | Separates SWERB_LOC_GPS geolocation points into X and Y coordinates for ease of maintenance. | SWERB_LOC_DATA, ADCODES |
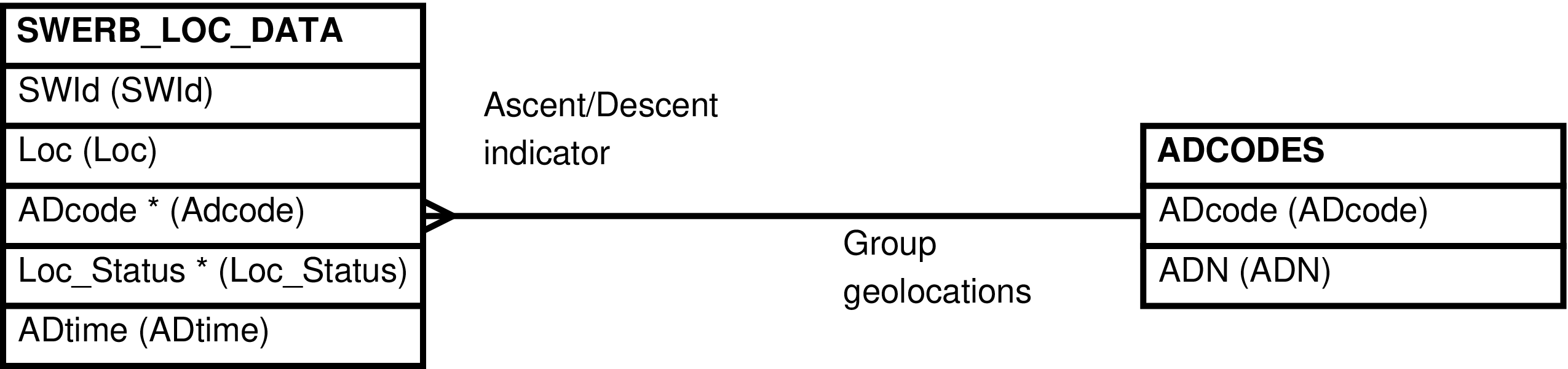
| SWERB_LOCS | SWERB_LOC_DATA row -- for every time a group is observed at a geolocated physical object, usually a grove or waterhole | Presents the relationship between the groups and physical features of the landscape in a more comprehensive manner for simpler querying. | SWERB_LOC_DATA, ADCODES |
| SWERB_UPLOAD | row uploaded into SWERB | This view returns no rows, it is used only to upload data into the swerb portion of Babase. | SWERB_DEPARTS_DATA, SWERB_DEPARTS_GPS, SWERB_BES, SWERB_DATA, SWERB_LOC_DATA |
| Weather Data | |||
| View | One row for each | Purpose | Tables/Views used |
| CURATED_TEMPS | WREADINGS row with a "curated" Tempmin or Tempmax; or for each 06:00 AM - 06:00 AM "day" having any rows in DIGITAL_WEATHER with a "curated" AirTemp_Min or AirTemp_Max | Collation of electronically and manually collected weather data into a single place. | WREADINGS TEMPMINS TEMPMAXS DIGITAL_WEATHER |
| MIN_MAXS | WREADINGS row | Analysis and correlation of manually collected weather data. | WREADINGS TEMPMINS TEMPMAXS RAINGAUGES |
| MIN_MAXS_SORTED | WREADINGS row | The MIN_MAXS view sorted for convienience. | WREADINGS TEMPMINS TEMPMAXS RAINGAUGES |
Table 4. The table_GRP Views
For information on the operations (INSERT, UPDATE, DELETE) allowed by each view and their actions on the underlying tables see The Babase Views of The Babase Reference Manual.
Figure 21. Query Defining the ACTOR_ACTEES View
WITH sampling AS (SELECT samples.date
, samples.observer
, ARRAY_AGG(DISTINCT members.grp) AS grps
FROM samples
JOIN stypes
ON stypes.stype = samples.stype
AND stypes.repr_interxns
JOIN members
ON members.sname = samples.sname
AND members.date = samples.date
WHERE samples.minsis > 0
GROUP BY samples.date, samples.observer)
SELECT interact_data.iid AS iid
, interact_data.sid AS sid
, interact_data.act AS act
, interact_data.date AS date
, interact_data.start AS start
, interact_data.stop AS stop
, interact_data.observer AS observer
, actor.partid AS actorid
, COALESCE(actor.sname, '998'::CHAR(3)) AS actor
, actorms.grp AS actor_grp
, actee.partid AS acteeid
, COALESCE(actee.sname, '998'::CHAR(3)) AS actee
, acteems.grp AS actee_grp
, interact_data.handwritten AS handwritten
, interact_data.exact_date AS exact_date
, COALESCE((actorms.grp = ANY(sampling.grps)
OR acteems.grp = ANY(sampling.grps))
, FALSE) AS repr_interxn
FROM interact_data
LEFT OUTER JOIN parts AS actor
ON (actor.iid = interact_data.iid AND actor.role = 'R')
LEFT OUTER JOIN parts AS actee
ON (actee.iid = interact_data.iid AND actee.role = 'E')
LEFT OUTER JOIN members AS actorms
ON (actorms.sname = actor.sname
AND actorms.date = interact_data.date)
LEFT OUTER JOIN members AS acteems
ON (acteems.sname = actee.sname
AND acteems.date = interact_data.date)
LEFT OUTER JOIN sampling
ON (sampling.date = interact_data.date
AND sampling.observer = interact_data.observer);
Figure 23. Query Defining the ANESTH_STATS View
SELECT anesths.dartid AS dartid
, count(*) AS ansamps
, avg(anesths.anamount) AS anamount_mean
, stddev(anesths.anamount) AS anamount_stddev
FROM anesths
GROUP BY anesths.dartid;
Figure 25. Query Defining the BODYTEMP_STATS View
SELECT bodytemps.dartid AS dartid
, count(*) AS btsamps
, avg(bodytemps.btemp) AS btemp_mean
, stddev(bodytemps.btemp) AS btemp_stddev
FROM bodytemps
GROUP BY bodytemps.dartid;
Figure 27. Query Defining the CENSUS_DEMOG View
SELECT census.cenid AS cenid
, census.sname AS sname
, census.date AS date
, census.grp AS grp
, census.status AS status
, census.cen AS cen
, demog.reference AS reference
, demog.comment AS comment
FROM census LEFT OUTER JOIN demog ON (census.cenid = demog.cenid);
Figure 29. Query Defining the CHEST_STATS View
SELECT chests.dartid AS dartid
, count(*) AS chsamps
, avg(chests.chcircum) AS chcircum_mean
, stddev(chests.chcircum) AS chcircum_stddev
, avg(chests.chunadjusted) AS chunadjusted_mean
, stddev(chests.chunadjusted) AS chunadjusted_stddev
FROM chests
GROUP BY chests.dartid;
Figure 31. Query Defining the CROWNRUMP_STATS View
SELECT crownrumps.dartid AS dartid
, count(*) AS crsamps
, avg(crownrumps.crlength) AS crlength_mean
, stddev(crownrumps.crlength) AS crlength_stddev
FROM crownrumps
GROUP BY crownrumps.dartid;
Figure 33. Query Defining the CYCLES_SEXSKINS View
SELECT cycles.cid AS cid
, cycles.sname AS sname
, cycles.seq AS seq
, cycles.series AS series
, sexskins.sxid AS sxid
, sexskins.date AS date
, sexskins.size AS size
, sexskins.color AS color
FROM cycles LEFT OUTER JOIN sexskins ON (cycles.cid = sexskins.cid);
Figure 35. Query Defining the CYCPOINTS_CYCLES View
SELECT cycles.cid AS cid
, cycles.sname AS sname
, cycles.seq AS seq
, cycles.series AS series
, cycpoints.cpid AS cpid
, cycpoints.date AS date
, cycpoints.edate AS edate
, cycpoints.ldate AS ldate
, cycpoints.code AS code
, cycpoints.source AS source
FROM cycles, cycpoints
WHERE cycles.cid = cycpoints.cid;
Figure 37. Query Defining the DSAMPLES View
SELECT dartings.dartid
, dartings.sname
, dartings.date
, members.grp
, blood_unspecs.num AS bloodunspec
, blood_paxgenes.num AS bloodpaxgene
, blood_purpletops.num AS bloodpurpletops
, blood_separators.num AS bloodseptube
, blood_cpts.num AS bloodcpt
, blood_trucultures.num AS bloodtruculture
, blood_smears.num AS bloodsmear
, tc_bloods.num AS tcblood
, hair_unspecs.num AS hairunspec
, hair_lengths.num AS hairlength
, hair_cu_zns.num AS haircu_zn
, teeth_3mouths.num AS mouthphotos3
, teeth_lmandmolds.num AS lmandmold
, teeth_lmaxmolds.num AS lmaxillamold
, teeth_lmol1mol2s.num AS lm1m2siliconemold
, skin_punchs.num AS skinpunch
, tc_skins.num AS tcskin
, vag_swabs.num AS vaginalswab
, cerv_swabs.num AS cervicalswab
, fecal_formalin.num AS fecal_formalin
, palm_swab.num AS palm_swab
, tongue_swab.num AS tongue_swab
, tooth_plaque_swab.num as tooth_plaque_swab
, vagswab_microbiome.num AS vagswab_microbiome
, glans_penis_swab.num AS glans_penis_swab
, fecal_microbiome.num AS fecal_microbiome
, nostrils_swab.num AS nostrils_swab
, skin_behind_ear_swab.num AS skin_behind_ear_swab
, skin_inside_elbow_swab.num AS skin_inside_elbow_swab
FROM dartings
JOIN members
ON dartings.sname = members.sname
AND dartings.date = members.date
LEFT JOIN dart_samples blood_unspecs
ON dartings.dartid = blood_unspecs.dartid
AND blood_unspecs.ds_type = 1
LEFT JOIN dart_samples blood_paxgenes
ON dartings.dartid = blood_paxgenes.dartid
AND blood_paxgenes.ds_type = 2
LEFT JOIN dart_samples blood_purpletops
ON dartings.dartid = blood_purpletops.dartid
AND blood_purpletops.ds_type = 3
LEFT JOIN dart_samples blood_separators
ON dartings.dartid = blood_separators.dartid
AND blood_separators.ds_type = 4
LEFT JOIN dart_samples blood_cpts
ON dartings.dartid = blood_cpts.dartid
AND blood_cpts.ds_type = 5
LEFT JOIN dart_samples blood_trucultures
ON dartings.dartid = blood_trucultures.dartid
AND blood_trucultures.ds_type = 6
LEFT JOIN dart_samples blood_smears
ON dartings.dartid = blood_smears.dartid
AND blood_smears.ds_type = 7
LEFT JOIN dart_samples hair_unspecs
ON dartings.dartid = hair_unspecs.dartid
AND hair_unspecs.ds_type = 8
LEFT JOIN dart_samples hair_lengths
ON dartings.dartid = hair_lengths.dartid
AND hair_lengths.ds_type = 9
LEFT JOIN dart_samples hair_cu_zns
ON dartings.dartid = hair_cu_zns.dartid
AND hair_cu_zns.ds_type = 10
LEFT JOIN dart_samples teeth_3mouths
ON dartings.dartid = teeth_3mouths.dartid
AND teeth_3mouths.ds_type = 11
LEFT JOIN dart_samples teeth_lmandmolds
ON dartings.dartid = teeth_lmandmolds.dartid
AND teeth_lmandmolds.ds_type = 12
LEFT JOIN dart_samples teeth_lmaxmolds
ON dartings.dartid = teeth_lmaxmolds.dartid
AND teeth_lmaxmolds.ds_type = 13
LEFT JOIN dart_samples teeth_lmol1mol2s
ON dartings.dartid = teeth_lmol1mol2s.dartid
AND teeth_lmol1mol2s.ds_type = 14
LEFT JOIN dart_samples skin_punchs
ON dartings.dartid = skin_punchs.dartid
AND skin_punchs.ds_type = 15
LEFT JOIN dart_samples vag_swabs
ON dartings.dartid = vag_swabs.dartid
AND vag_swabs.ds_type = 16
LEFT JOIN dart_samples cerv_swabs
ON dartings.dartid = cerv_swabs.dartid
AND cerv_swabs.ds_type = 17
LEFT JOIN dart_samples tc_bloods
ON dartings.dartid = tc_bloods.dartid
AND tc_bloods.ds_type = 18
LEFT JOIN dart_samples tc_skins
ON dartings.dartid = tc_skins.dartid
AND tc_skins.ds_type = 19
LEFT JOIN dart_samples fecal_formalin
ON dartings.dartid = fecal_formalin.dartid
AND fecal_formalin.ds_type = 20
LEFT JOIN dart_samples palm_swab
ON dartings.dartid = palm_swab.dartid
AND palm_swab.ds_type = 22
LEFT JOIN dart_samples tongue_swab
ON dartings.dartid = tongue_swab.dartid
AND tongue_swab.ds_type = 23
LEFT JOIN dart_samples tooth_plaque_swab
ON dartings.dartid = tooth_plaque_swab.dartid
AND tooth_plaque_swab.ds_type = 24
LEFT JOIN dart_samples vagswab_microbiome
ON dartings.dartid = vagswab_microbiome.dartid
AND vagswab_microbiome.ds_type = 25
LEFT JOIN dart_samples glans_penis_swab
ON dartings.dartid = glans_penis_swab.dartid
AND glans_penis_swab.ds_type = 26
LEFT JOIN dart_samples fecal_microbiome
ON dartings.dartid = fecal_microbiome.dartid
AND fecal_microbiome.ds_type = 27
LEFT JOIN dart_samples nostrils_swab
ON dartings.dartid = nostrils_swab.dartid
AND nostrils_swab.ds_type = 28
LEFT JOIN dart_samples skin_behind_ear_swab
ON dartings.dartid = skin_behind_ear_swab.dartid
AND skin_behind_ear_swab.ds_type = 29
LEFT JOIN dart_samples skin_inside_elbow_swab
ON dartings.dartid = skin_inside_elbow_swab.dartid
AND skin_inside_elbow_swab.ds_type = 30;
Figure 38. Query Defining the DEMOG_CENSUS View
SELECT census.cenid AS cenid
, census.sname AS sname
, census.date AS date
, census.grp AS grp
, census.status AS status
, census.cen AS cen
, demog.reference AS reference
, demog.comment AS comment
FROM census, demog
WHERE census.cenid = demog.cenid;
Figure 40. Query Defining the DENT_CODES View
SELECT teethdartids.dartid AS dartid
, rum3.rum3tstate AS rum3tstate
, rum3.rum3tcondition AS rum3tcondition
, rum2.rum2tstate AS rum2tstate
, rum2.rum2tcondition AS rum2tcondition
, rum1.rum1tstate AS rum1tstate
, rum1.rum1tcondition AS rum1tcondition
, rup2.rup2tstate AS rup2tstate
, rup2.rup2tcondition AS rup2tcondition
, rup1.rup1tstate AS rup1tstate
, rup1.rup1tcondition AS rup1tcondition
, ruc.ructstate AS ructstate
, ruc.ructcondition AS ructcondition
, rui2.rui2tstate AS rui2tstate
, rui2.rui2tcondition AS rui2tcondition
, rui1.rui1tstate AS rui1tstate
, rui1.rui1tcondition AS rui1tcondition
, lui1.lui1tstate AS lui1tstate
, lui1.lui1tcondition AS lui1tcondition
, lui2.lui2tstate AS lui2tstate
, lui2.lui2tcondition AS lui2tcondition
, luc.luctstate AS luctstate
, luc.luctcondition AS luctcondition
, lup1.lup1tstate AS lup1tstate
, lup1.lup1tcondition AS lup1tcondition
, lup2.lup2tstate AS lup2tstate
, lup2.lup2tcondition AS lup2tcondition
, lum1.lum1tstate AS lum1tstate
, lum1.lum1tcondition AS lum1tcondition
, lum2.lum2tstate AS lum2tstate
, lum2.lum2tcondition AS lum2tcondition
, lum3.lum3tstate AS lum3tstate
, lum3.lum3tcondition AS lum3tcondition
, llm3.llm3tstate AS llm3tstate
, llm3.llm3tcondition AS llm3tcondition
, llm2.llm2tstate AS llm2tstate
, llm2.llm2tcondition AS llm2tcondition
, llm1.llm1tstate AS llm1tstate
, llm1.llm1tcondition AS llm1tcondition
, llp2.llp2tstate AS llp2tstate
, llp2.llp2tcondition AS llp2tcondition
, llp1.llp1tstate AS llp1tstate
, llp1.llp1tcondition AS llp1tcondition
, llc.llctstate AS llctstate
, llc.llctcondition AS llctcondition
, lli2.lli2tstate AS lli2tstate
, lli2.lli2tcondition AS lli2tcondition
, lli1.lli1tstate AS lli1tstate
, lli1.lli1tcondition AS lli1tcondition
, rli1.rli1tstate AS rli1tstate
, rli1.rli1tcondition AS rli1tcondition
, rli2.rli2tstate AS rli2tstate
, rli2.rli2tcondition AS rli2tcondition
, rlc.rlctstate AS rlctstate
, rlc.rlctcondition AS rlctcondition
, rlp1.rlp1tstate AS rlp1tstate
, rlp1.rlp1tcondition AS rlp1tcondition
, rlp2.rlp2tstate AS rlp2tstate
, rlp2.rlp2tcondition AS rlp2tcondition
, rlm1.rlm1tstate AS rlm1tstate
, rlm1.rlm1tcondition AS rlm1tcondition
, rlm2.rlm2tstate AS rlm2tstate
, rlm2.rlm2tcondition AS rlm2tcondition
, rlm3.rlm3tstate AS rlm3tstate
, rlm3.rlm3tcondition AS rlm3tcondition
, drum2.drum2tstate AS drum2tstate
, drum2.drum2tcondition AS drum2tcondition
, drum1.drum1tstate AS drum1tstate
, drum1.drum1tcondition AS drum1tcondition
, druc.dructstate AS dructstate
, druc.dructcondition AS dructcondition
, drui2.drui2tstate AS drui2tstate
, drui2.drui2tcondition AS drui2tcondition
, drui1.drui1tstate AS drui1tstate
, drui1.drui1tcondition AS drui1tcondition
, dlui1.dlui1tstate AS dlui1tstate
, dlui1.dlui1tcondition AS dlui1tcondition
, dlui2.dlui2tstate AS dlui2tstate
, dlui2.dlui2tcondition AS dlui2tcondition
, dluc.dluctstate AS dluctstate
, dluc.dluctcondition AS dluctcondition
, dlum1.dlum1tstate AS dlum1tstate
, dlum1.dlum1tcondition AS dlum1tcondition
, dlum2.dlum2tstate AS dlum2tstate
, dlum2.dlum2tcondition AS dlum2tcondition
, dllm2.dllm2tstate AS dllm2tstate
, dllm2.dllm2tcondition AS dllm2tcondition
, dllm1.dllm1tstate AS dllm1tstate
, dllm1.dllm1tcondition AS dllm1tcondition
, dllc.dllctstate AS dllctstate
, dllc.dllctcondition AS dllctcondition
, dlli2.dlli2tstate AS dlli2tstate
, dlli2.dlli2tcondition AS dlli2tcondition
, dlli1.dlli1tstate AS dlli1tstate
, dlli1.dlli1tcondition AS dlli1tcondition
, drli1.drli1tstate AS drli1tstate
, drli1.drli1tcondition AS drli1tcondition
, drli2.drli2tstate AS drli2tstate
, drli2.drli2tcondition AS drli2tcondition
, drlc.drlctstate AS drlctstate
, drlc.drlctcondition AS drlctcondition
, drlm1.drlm1tstate AS drlm1tstate
, drlm1.drlm1tcondition AS drlm1tcondition
, drlm2.drlm2tstate AS drlm2tstate
, drlm2.drlm2tcondition AS drlm2tcondition
FROM (SELECT teeth.dartid
FROM teeth
GROUP BY teeth.dartid)
AS teethdartids
LEFT OUTER JOIN
(SELECT teeth.dartid AS rum3dartid
, teeth.tstate AS rum3tstate
, teeth.tcondition AS rum3tcondition
FROM teeth
WHERE teeth.tooth = 'rum3')
AS rum3
ON rum3.rum3dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rum2dartid
, teeth.tstate AS rum2tstate
, teeth.tcondition AS rum2tcondition
FROM teeth
WHERE teeth.tooth = 'rum2')
AS rum2
ON rum2.rum2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rum1dartid
, teeth.tstate AS rum1tstate
, teeth.tcondition AS rum1tcondition
FROM teeth
WHERE teeth.tooth = 'rum1')
AS rum1
ON rum1.rum1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rup2dartid
, teeth.tstate AS rup2tstate
, teeth.tcondition AS rup2tcondition
FROM teeth
WHERE teeth.tooth = 'rup2')
AS rup2
ON rup2.rup2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rup1dartid
, teeth.tstate AS rup1tstate
, teeth.tcondition AS rup1tcondition
FROM teeth
WHERE teeth.tooth = 'rup1')
AS rup1
ON rup1.rup1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rucdartid
, teeth.tstate AS ructstate
, teeth.tcondition AS ructcondition
FROM teeth
WHERE teeth.tooth = 'ruc')
AS ruc
ON ruc.rucdartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rui2dartid
, teeth.tstate AS rui2tstate
, teeth.tcondition AS rui2tcondition
FROM teeth
WHERE teeth.tooth = 'rui2')
AS rui2
ON rui2.rui2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rui1dartid
, teeth.tstate AS rui1tstate
, teeth.tcondition AS rui1tcondition
FROM teeth
WHERE teeth.tooth = 'rui1')
AS rui1
ON rui1.rui1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS lui1dartid
, teeth.tstate AS lui1tstate
, teeth.tcondition AS lui1tcondition
FROM teeth
WHERE teeth.tooth = 'lui1')
AS lui1
ON lui1.lui1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS lui2dartid
, teeth.tstate AS lui2tstate
, teeth.tcondition AS lui2tcondition
FROM teeth
WHERE teeth.tooth = 'lui2')
AS lui2
ON lui2.lui2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS lucdartid
, teeth.tstate AS luctstate
, teeth.tcondition AS luctcondition
FROM teeth
WHERE teeth.tooth = 'luc')
AS luc
ON luc.lucdartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS lup1dartid
, teeth.tstate AS lup1tstate
, teeth.tcondition AS lup1tcondition
FROM teeth
WHERE teeth.tooth = 'lup1')
AS lup1
ON lup1.lup1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS lup2dartid
, teeth.tstate AS lup2tstate
, teeth.tcondition AS lup2tcondition
FROM teeth
WHERE teeth.tooth = 'lup2')
AS lup2
ON lup2.lup2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS lum1dartid
, teeth.tstate AS lum1tstate
, teeth.tcondition AS lum1tcondition
FROM teeth
WHERE teeth.tooth = 'lum1')
AS lum1
ON lum1.lum1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS lum2dartid
, teeth.tstate AS lum2tstate
, teeth.tcondition AS lum2tcondition
FROM teeth
WHERE teeth.tooth = 'lum2')
AS lum2
ON lum2.lum2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS lum3dartid
, teeth.tstate AS lum3tstate
, teeth.tcondition AS lum3tcondition
FROM teeth
WHERE teeth.tooth = 'lum3')
AS lum3
ON lum3.lum3dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS llm3dartid
, teeth.tstate AS llm3tstate
, teeth.tcondition AS llm3tcondition
FROM teeth
WHERE teeth.tooth = 'llm3')
AS llm3
ON llm3.llm3dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS llm2dartid
, teeth.tstate AS llm2tstate
, teeth.tcondition AS llm2tcondition
FROM teeth
WHERE teeth.tooth = 'llm2')
AS llm2
ON llm2.llm2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS llm1dartid
, teeth.tstate AS llm1tstate
, teeth.tcondition AS llm1tcondition
FROM teeth
WHERE teeth.tooth = 'llm1')
AS llm1
ON llm1.llm1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS llp2dartid
, teeth.tstate AS llp2tstate
, teeth.tcondition AS llp2tcondition
FROM teeth
WHERE teeth.tooth = 'llp2')
AS llp2
ON llp2.llp2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS llp1dartid
, teeth.tstate AS llp1tstate
, teeth.tcondition AS llp1tcondition
FROM teeth
WHERE teeth.tooth = 'llp1')
AS llp1
ON llp1.llp1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS llcdartid
, teeth.tstate AS llctstate
, teeth.tcondition AS llctcondition
FROM teeth
WHERE teeth.tooth = 'llc')
AS llc
ON llc.llcdartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS lli2dartid
, teeth.tstate AS lli2tstate
, teeth.tcondition AS lli2tcondition
FROM teeth
WHERE teeth.tooth = 'lli2')
AS lli2
ON lli2.lli2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS lli1dartid
, teeth.tstate AS lli1tstate
, teeth.tcondition AS lli1tcondition
FROM teeth
WHERE teeth.tooth = 'lli1')
AS lli1
ON lli1.lli1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rli1dartid
, teeth.tstate AS rli1tstate
, teeth.tcondition AS rli1tcondition
FROM teeth
WHERE teeth.tooth = 'rli1')
AS rli1
ON rli1.rli1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rli2dartid
, teeth.tstate AS rli2tstate
, teeth.tcondition AS rli2tcondition
FROM teeth
WHERE teeth.tooth = 'rli2')
AS rli2
ON rli2.rli2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rlcdartid
, teeth.tstate AS rlctstate
, teeth.tcondition AS rlctcondition
FROM teeth
WHERE teeth.tooth = 'rlc')
AS rlc
ON rlc.rlcdartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rlp1dartid
, teeth.tstate AS rlp1tstate
, teeth.tcondition AS rlp1tcondition
FROM teeth
WHERE teeth.tooth = 'rlp1')
AS rlp1
ON rlp1.rlp1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rlp2dartid
, teeth.tstate AS rlp2tstate
, teeth.tcondition AS rlp2tcondition
FROM teeth
WHERE teeth.tooth = 'rlp2')
AS rlp2
ON rlp2.rlp2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rlm1dartid
, teeth.tstate AS rlm1tstate
, teeth.tcondition AS rlm1tcondition
FROM teeth
WHERE teeth.tooth = 'rlm1')
AS rlm1
ON rlm1.rlm1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rlm2dartid
, teeth.tstate AS rlm2tstate
, teeth.tcondition AS rlm2tcondition
FROM teeth
WHERE teeth.tooth = 'rlm2')
AS rlm2
ON rlm2.rlm2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS rlm3dartid
, teeth.tstate AS rlm3tstate
, teeth.tcondition AS rlm3tcondition
FROM teeth
WHERE teeth.tooth = 'rlm3')
AS rlm3
ON rlm3.rlm3dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS drum2dartid
, teeth.tstate AS drum2tstate
, teeth.tcondition AS drum2tcondition
FROM teeth
WHERE teeth.tooth = 'drum2')
AS drum2
ON drum2.drum2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS drum1dartid
, teeth.tstate AS drum1tstate
, teeth.tcondition AS drum1tcondition
FROM teeth
WHERE teeth.tooth = 'drum1')
AS drum1
ON drum1.drum1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS drucdartid
, teeth.tstate AS dructstate
, teeth.tcondition AS dructcondition
FROM teeth
WHERE teeth.tooth = 'druc')
AS druc
ON druc.drucdartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS drui2dartid
, teeth.tstate AS drui2tstate
, teeth.tcondition AS drui2tcondition
FROM teeth
WHERE teeth.tooth = 'drui2')
AS drui2
ON drui2.drui2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS drui1dartid
, teeth.tstate AS drui1tstate
, teeth.tcondition AS drui1tcondition
FROM teeth
WHERE teeth.tooth = 'drui1')
AS drui1
ON drui1.drui1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS dlui1dartid
, teeth.tstate AS dlui1tstate
, teeth.tcondition AS dlui1tcondition
FROM teeth
WHERE teeth.tooth = 'dlui1')
AS dlui1
ON dlui1.dlui1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS dlui2dartid
, teeth.tstate AS dlui2tstate
, teeth.tcondition AS dlui2tcondition
FROM teeth
WHERE teeth.tooth = 'dlui2')
AS dlui2
ON dlui2.dlui2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS dlucdartid
, teeth.tstate AS dluctstate
, teeth.tcondition AS dluctcondition
FROM teeth
WHERE teeth.tooth = 'dluc')
AS dluc
ON dluc.dlucdartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS dlum1dartid
, teeth.tstate AS dlum1tstate
, teeth.tcondition AS dlum1tcondition
FROM teeth
WHERE teeth.tooth = 'dlum1')
AS dlum1
ON dlum1.dlum1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS dlum2dartid
, teeth.tstate AS dlum2tstate
, teeth.tcondition AS dlum2tcondition
FROM teeth
WHERE teeth.tooth = 'dlum2')
AS dlum2
ON dlum2.dlum2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS dllm2dartid
, teeth.tstate AS dllm2tstate
, teeth.tcondition AS dllm2tcondition
FROM teeth
WHERE teeth.tooth = 'dllm2')
AS dllm2
ON dllm2.dllm2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS dllm1dartid
, teeth.tstate AS dllm1tstate
, teeth.tcondition AS dllm1tcondition
FROM teeth
WHERE teeth.tooth = 'dllm1')
AS dllm1
ON dllm1.dllm1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS dllcdartid
, teeth.tstate AS dllctstate
, teeth.tcondition AS dllctcondition
FROM teeth
WHERE teeth.tooth = 'dllc')
AS dllc
ON dllc.dllcdartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS dlli2dartid
, teeth.tstate AS dlli2tstate
, teeth.tcondition AS dlli2tcondition
FROM teeth
WHERE teeth.tooth = 'dlli2')
AS dlli2
ON dlli2.dlli2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS dlli1dartid
, teeth.tstate AS dlli1tstate
, teeth.tcondition AS dlli1tcondition
FROM teeth
WHERE teeth.tooth = 'dlli1')
AS dlli1
ON dlli1.dlli1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS drli1dartid
, teeth.tstate AS drli1tstate
, teeth.tcondition AS drli1tcondition
FROM teeth
WHERE teeth.tooth = 'drli1')
AS drli1
ON drli1.drli1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS drli2dartid
, teeth.tstate AS drli2tstate
, teeth.tcondition AS drli2tcondition
FROM teeth
WHERE teeth.tooth = 'drli2')
AS drli2
ON drli2.drli2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS drlcdartid
, teeth.tstate AS drlctstate
, teeth.tcondition AS drlctcondition
FROM teeth
WHERE teeth.tooth = 'drlc')
AS drlc
ON drlc.drlcdartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS drlm1dartid
, teeth.tstate AS drlm1tstate
, teeth.tcondition AS drlm1tcondition
FROM teeth
WHERE teeth.tooth = 'drlm1')
AS drlm1
ON drlm1.drlm1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS drlm2dartid
, teeth.tstate AS drlm2tstate
, teeth.tcondition AS drlm2tcondition
FROM teeth
WHERE teeth.tooth = 'drlm2')
AS drlm2
ON drlm2.drlm2dartid = teethdartids.dartid;
Figure 42. Query Defining the DENT_SITES View
SELECT teethdartids.dartid AS dartid
, s1.s1tstate AS s1tstate
, s1.s1tcondition AS s1tcondition
, s1.s1deciduous AS s1deciduous
, s2.s2tstate AS s2tstate
, s2.s2tcondition AS s2tcondition
, s2.s2deciduous AS s2deciduous
, s3.s3tstate AS s3tstate
, s3.s3tcondition AS s3tcondition
, s3.s3deciduous AS s3deciduous
, s4.s4tstate AS s4tstate
, s4.s4tcondition AS s4tcondition
, s4.s4deciduous AS s4deciduous
, s5.s5tstate AS s5tstate
, s5.s5tcondition AS s5tcondition
, s5.s5deciduous AS s5deciduous
, s6.s6tstate AS s6tstate
, s6.s6tcondition AS s6tcondition
, s6.s6deciduous AS s6deciduous
, s7.s7tstate AS s7tstate
, s7.s7tcondition AS s7tcondition
, s7.s7deciduous AS s7deciduous
, s8.s8tstate AS s8tstate
, s8.s8tcondition AS s8tcondition
, s8.s8deciduous AS s8deciduous
, s9.s9tstate AS s9tstate
, s9.s9tcondition AS s9tcondition
, s9.s9deciduous AS s9deciduous
, s10.s10tstate AS s10tstate
, s10.s10tcondition AS s10tcondition
, s10.s10deciduous AS s10deciduous
, s11.s11tstate AS s11tstate
, s11.s11tcondition AS s11tcondition
, s11.s11deciduous AS s11deciduous
, s12.s12tstate AS s12tstate
, s12.s12tcondition AS s12tcondition
, s12.s12deciduous AS s12deciduous
, s13.s13tstate AS s13tstate
, s13.s13tcondition AS s13tcondition
, s13.s13deciduous AS s13deciduous
, s14.s14tstate AS s14tstate
, s14.s14tcondition AS s14tcondition
, s14.s14deciduous AS s14deciduous
, s15.s15tstate AS s15tstate
, s15.s15tcondition AS s15tcondition
, s15.s15deciduous AS s15deciduous
, s16.s16tstate AS s16tstate
, s16.s16tcondition AS s16tcondition
, s16.s16deciduous AS s16deciduous
, s17.s17tstate AS s17tstate
, s17.s17tcondition AS s17tcondition
, s17.s17deciduous AS s17deciduous
, s18.s18tstate AS s18tstate
, s18.s18tcondition AS s18tcondition
, s18.s18deciduous AS s18deciduous
, s19.s19tstate AS s19tstate
, s19.s19tcondition AS s19tcondition
, s19.s19deciduous AS s19deciduous
, s20.s20tstate AS s20tstate
, s20.s20tcondition AS s20tcondition
, s20.s20deciduous AS s20deciduous
, s21.s21tstate AS s21tstate
, s21.s21tcondition AS s21tcondition
, s21.s21deciduous AS s21deciduous
, s22.s22tstate AS s22tstate
, s22.s22tcondition AS s22tcondition
, s22.s22deciduous AS s22deciduous
, s23.s23tstate AS s23tstate
, s23.s23tcondition AS s23tcondition
, s23.s23deciduous AS s23deciduous
, s24.s24tstate AS s24tstate
, s24.s24tcondition AS s24tcondition
, s24.s24deciduous AS s24deciduous
, s25.s25tstate AS s25tstate
, s25.s25tcondition AS s25tcondition
, s25.s25deciduous AS s25deciduous
, s26.s26tstate AS s26tstate
, s26.s26tcondition AS s26tcondition
, s26.s26deciduous AS s26deciduous
, s27.s27tstate AS s27tstate
, s27.s27tcondition AS s27tcondition
, s27.s27deciduous AS s27deciduous
, s28.s28tstate AS s28tstate
, s28.s28tcondition AS s28tcondition
, s28.s28deciduous AS s28deciduous
, s29.s29tstate AS s29tstate
, s29.s29tcondition AS s29tcondition
, s29.s29deciduous AS s29deciduous
, s30.s30tstate AS s30tstate
, s30.s30tcondition AS s30tcondition
, s30.s30deciduous AS s30deciduous
, s31.s31tstate AS s31tstate
, s31.s31tcondition AS s31tcondition
, s31.s31deciduous AS s31deciduous
, s32.s32tstate AS s32tstate
, s32.s32tcondition AS s32tcondition
, s32.s32deciduous AS s32deciduous
FROM (SELECT teeth.dartid
FROM teeth
GROUP BY teeth.dartid)
AS teethdartids
LEFT OUTER JOIN
(SELECT teeth.dartid AS s1dartid
, teeth.tstate AS s1tstate
, teeth.tcondition AS s1tcondition
, toothcodes.deciduous AS s1deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '1'
AND teeth.tooth = toothcodes.tooth)
AS s1
ON s1.s1dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s2dartid
, teeth.tstate AS s2tstate
, teeth.tcondition AS s2tcondition
, toothcodes.deciduous AS s2deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '2'
AND teeth.tooth = toothcodes.tooth)
AS s2
ON s2.s2dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s3dartid
, teeth.tstate AS s3tstate
, teeth.tcondition AS s3tcondition
, toothcodes.deciduous AS s3deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '3'
AND teeth.tooth = toothcodes.tooth)
AS s3
ON s3.s3dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s4dartid
, teeth.tstate AS s4tstate
, teeth.tcondition AS s4tcondition
, toothcodes.deciduous AS s4deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '4'
AND teeth.tooth = toothcodes.tooth)
AS s4
ON s4.s4dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s5dartid
, teeth.tstate AS s5tstate
, teeth.tcondition AS s5tcondition
, toothcodes.deciduous AS s5deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '5'
AND teeth.tooth = toothcodes.tooth)
AS s5
ON s5.s5dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s6dartid
, teeth.tstate AS s6tstate
, teeth.tcondition AS s6tcondition
, toothcodes.deciduous AS s6deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '6'
AND teeth.tooth = toothcodes.tooth)
AS s6
ON s6.s6dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s7dartid
, teeth.tstate AS s7tstate
, teeth.tcondition AS s7tcondition
, toothcodes.deciduous AS s7deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '7'
AND teeth.tooth = toothcodes.tooth)
AS s7
ON s7.s7dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s8dartid
, teeth.tstate AS s8tstate
, teeth.tcondition AS s8tcondition
, toothcodes.deciduous AS s8deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '8'
AND teeth.tooth = toothcodes.tooth)
AS s8
ON s8.s8dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s9dartid
, teeth.tstate AS s9tstate
, teeth.tcondition AS s9tcondition
, toothcodes.deciduous AS s9deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '9'
AND teeth.tooth = toothcodes.tooth)
AS s9
ON s9.s9dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s10dartid
, teeth.tstate AS s10tstate
, teeth.tcondition AS s10tcondition
, toothcodes.deciduous AS s10deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '10'
AND teeth.tooth = toothcodes.tooth)
AS s10
ON s10.s10dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s11dartid
, teeth.tstate AS s11tstate
, teeth.tcondition AS s11tcondition
, toothcodes.deciduous AS s11deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '11'
AND teeth.tooth = toothcodes.tooth)
AS s11
ON s11.s11dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s12dartid
, teeth.tstate AS s12tstate
, teeth.tcondition AS s12tcondition
, toothcodes.deciduous AS s12deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '12'
AND teeth.tooth = toothcodes.tooth)
AS s12
ON s12.s12dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s13dartid
, teeth.tstate AS s13tstate
, teeth.tcondition AS s13tcondition
, toothcodes.deciduous AS s13deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '13'
AND teeth.tooth = toothcodes.tooth)
AS s13
ON s13.s13dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s14dartid
, teeth.tstate AS s14tstate
, teeth.tcondition AS s14tcondition
, toothcodes.deciduous AS s14deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '14'
AND teeth.tooth = toothcodes.tooth)
AS s14
ON s14.s14dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s15dartid
, teeth.tstate AS s15tstate
, teeth.tcondition AS s15tcondition
, toothcodes.deciduous AS s15deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '15'
AND teeth.tooth = toothcodes.tooth)
AS s15
ON s15.s15dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s16dartid
, teeth.tstate AS s16tstate
, teeth.tcondition AS s16tcondition
, toothcodes.deciduous AS s16deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '16'
AND teeth.tooth = toothcodes.tooth)
AS s16
ON s16.s16dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s17dartid
, teeth.tstate AS s17tstate
, teeth.tcondition AS s17tcondition
, toothcodes.deciduous AS s17deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '17'
AND teeth.tooth = toothcodes.tooth)
AS s17
ON s17.s17dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s18dartid
, teeth.tstate AS s18tstate
, teeth.tcondition AS s18tcondition
, toothcodes.deciduous AS s18deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '18'
AND teeth.tooth = toothcodes.tooth)
AS s18
ON s18.s18dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s19dartid
, teeth.tstate AS s19tstate
, teeth.tcondition AS s19tcondition
, toothcodes.deciduous AS s19deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '19'
AND teeth.tooth = toothcodes.tooth)
AS s19
ON s19.s19dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s20dartid
, teeth.tstate AS s20tstate
, teeth.tcondition AS s20tcondition
, toothcodes.deciduous AS s20deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '20'
AND teeth.tooth = toothcodes.tooth)
AS s20
ON s20.s20dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s21dartid
, teeth.tstate AS s21tstate
, teeth.tcondition AS s21tcondition
, toothcodes.deciduous AS s21deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '21'
AND teeth.tooth = toothcodes.tooth)
AS s21
ON s21.s21dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s22dartid
, teeth.tstate AS s22tstate
, teeth.tcondition AS s22tcondition
, toothcodes.deciduous AS s22deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '22'
AND teeth.tooth = toothcodes.tooth)
AS s22
ON s22.s22dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s23dartid
, teeth.tstate AS s23tstate
, teeth.tcondition AS s23tcondition
, toothcodes.deciduous AS s23deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '23'
AND teeth.tooth = toothcodes.tooth)
AS s23
ON s23.s23dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s24dartid
, teeth.tstate AS s24tstate
, teeth.tcondition AS s24tcondition
, toothcodes.deciduous AS s24deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '24'
AND teeth.tooth = toothcodes.tooth)
AS s24
ON s24.s24dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s25dartid
, teeth.tstate AS s25tstate
, teeth.tcondition AS s25tcondition
, toothcodes.deciduous AS s25deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '25'
AND teeth.tooth = toothcodes.tooth)
AS s25
ON s25.s25dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s26dartid
, teeth.tstate AS s26tstate
, teeth.tcondition AS s26tcondition
, toothcodes.deciduous AS s26deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '26'
AND teeth.tooth = toothcodes.tooth)
AS s26
ON s26.s26dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s27dartid
, teeth.tstate AS s27tstate
, teeth.tcondition AS s27tcondition
, toothcodes.deciduous AS s27deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '27'
AND teeth.tooth = toothcodes.tooth)
AS s27
ON s27.s27dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s28dartid
, teeth.tstate AS s28tstate
, teeth.tcondition AS s28tcondition
, toothcodes.deciduous AS s28deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '28'
AND teeth.tooth = toothcodes.tooth)
AS s28
ON s28.s28dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s29dartid
, teeth.tstate AS s29tstate
, teeth.tcondition AS s29tcondition
, toothcodes.deciduous AS s29deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '29'
AND teeth.tooth = toothcodes.tooth)
AS s29
ON s29.s29dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s30dartid
, teeth.tstate AS s30tstate
, teeth.tcondition AS s30tcondition
, toothcodes.deciduous AS s30deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '30'
AND teeth.tooth = toothcodes.tooth)
AS s30
ON s30.s30dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s31dartid
, teeth.tstate AS s31tstate
, teeth.tcondition AS s31tcondition
, toothcodes.deciduous AS s31deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '31'
AND teeth.tooth = toothcodes.tooth)
AS s31
ON s31.s31dartid = teethdartids.dartid
LEFT OUTER JOIN
(SELECT teeth.dartid AS s32dartid
, teeth.tstate AS s32tstate
, teeth.tcondition AS s32tcondition
, toothcodes.deciduous AS s32deciduous
FROM toothcodes, teeth
WHERE toothcodes.toothsite = '32'
AND teeth.tooth = toothcodes.tooth)
AS s32
ON s32.s32dartid = teethdartids.dartid;
Figure 44. Query Defining the ESTROGENS View
SELECT hormone_sample_data.tid
, hormone_prep_series.hpsid
, hormone_result_data.hrid
, hormone_sample_data.hsid
, biograph.sname
, tissue_data.collection_date
, tissue_data.collection_date_status AS collection_date_status
, hormone_sample_data.fzdried_date AS fzdried_date
, hormone_sample_data.sifted_date AS sifted_date
, meoh_ext.procedure_date AS me_extracted
, spe.procedure_date AS sp_extracted
, hormone_result_data.raw_ng_g AS raw_ng_g
, corrected_hormone(hormone_result_data.raw_ng_g, hormone_kits.correction) AS corrected_ng_g
, hormone_result_data.assay_date
, hormone_kits.hormone AS hormone
, hormone_result_data.kit AS kit
, hormone_sample_data.comments AS sample_comments
, hormone_result_data.comments AS result_comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ
JOIN hormone_prep_series
ON hormone_prep_series.tid = hormone_sample_data.tid
JOIN hormone_result_data
ON hormone_result_data.hpsid = hormone_prep_series.hpsid
JOIN hormone_kits
ON hormone_kits.kit = hormone_result_data.kit
AND hormone_kits.correction IS NOT NULL
AND hormone_kits.hormone = 'E'
LEFT JOIN hormone_prep_data AS meoh_ext
ON meoh_ext.procedure = 'MEOH_EXT'
AND meoh_ext.hpsid = hormone_prep_series.hpsid
LEFT JOIN hormone_prep_data AS spe
ON spe.procedure = 'SPE'
AND spe.hpsid = hormone_prep_series.hpsid;
Figure 46. Query Defining the GLUCOCORTICOIDS View
SELECT hormone_sample_data.tid
, hormone_prep_series.hpsid
, hormone_result_data.hrid
, hormone_sample_data.hsid
, biograph.sname
, tissue_data.collection_date
, tissue_data.collection_date_status AS collection_date_status
, hormone_sample_data.fzdried_date AS fzdried_date
, hormone_sample_data.sifted_date AS sifted_date
, meoh_ext.procedure_date AS me_extracted
, spe.procedure_date AS sp_extracted
, hormone_result_data.raw_ng_g AS raw_ng_g
, corrected_hormone(hormone_result_data.raw_ng_g, hormone_kits.correction) AS corrected_ng_g
, hormone_result_data.assay_date
, hormone_kits.hormone AS hormone
, hormone_result_data.kit AS kit
, hormone_sample_data.comments AS sample_comments
, hormone_result_data.comments AS result_comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ
JOIN hormone_prep_series
ON hormone_prep_series.tid = hormone_sample_data.tid
JOIN hormone_result_data
ON hormone_result_data.hpsid = hormone_prep_series.hpsid
JOIN hormone_kits
ON hormone_kits.kit = hormone_result_data.kit
AND hormone_kits.correction IS NOT NULL
AND hormone_kits.hormone = 'GC'
LEFT JOIN hormone_prep_data AS meoh_ext
ON meoh_ext.procedure = 'MEOH_EXT'
AND meoh_ext.hpsid = hormone_prep_series.hpsid
LEFT JOIN hormone_prep_data AS spe
ON spe.procedure = 'SPE'
AND spe.hpsid = hormone_prep_series.hpsid;
Figure 48. Query Defining the GROUPS_HISTORY View
SELECT groups.gid AS gid
, groups.name AS name
, groups.from_group AS from_group
, groups.to_group AS to_group
, CASE
WHEN groups.from_group IS NULL
AND NOT EXISTS (SELECT 1
FROM groups AS from_groups
WHERE from_groups.to_group = groups.gid)
THEN groups.permanent
ELSE groups.start
END AS first_observed
, CASE
WHEN groups.study_grp IS NULL
THEN NULL
WHEN groups.from_group IS NULL
AND NOT EXISTS (SELECT 1
FROM groups AS from_groups
WHERE from_groups.to_group = groups.gid)
THEN groups.permanent
ELSE (SELECT date
FROM census
WHERE census.grp = groups.gid
AND census.cen
ORDER BY date
LIMIT 1)
END AS first_study_grp_census
, groups.permanent AS permanent
, (SELECT descgroups_start.start
FROM babase.groups AS descgroups_start
WHERE descgroups_start.from_group = groups.gid
OR descgroups_start.gid = groups.to_group
ORDER BY descgroups_start.start
LIMIT 1
) AS impermanent
, groups.cease_to_exist AS cease_to_exist
, groups.last_reg_census AS last_reg_census
, groups.study_grp
FROM babase.groups;
Figure 50. Query Defining the HORMONE_PREPS View
SELECT hormone_sample_data.tid AS tid
, hormone_sample_data.hsid AS hsid
, unique_indivs.individ AS individ
, biograph.sname AS sname
, hormone_prep_series.hpsid AS hpsid
, hormone_prep_series.series AS series
, hormone_prep_data.hpid AS hpid
, hormone_prep_data.procedure AS procedure
, hormone_prep_data.procedure_date AS procedure_date
, hormone_prep_data.comments AS comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ
JOIN hormone_prep_series
ON hormone_prep_series.tid = hormone_sample_data.tid
JOIN hormone_prep_data
ON hormone_prep_data.hpsid = hormone_prep_series.hpsid;
Figure 52. Query Defining the HORMONE_RESULTS View
SELECT hormone_sample_data.tid AS tid
, hormone_sample_data.hsid AS hsid
, unique_indivs.individ AS individ
, biograph.sname AS sname
, hormone_prep_series.hpsid AS hpsid
, hormone_prep_series.series AS series
, hormone_result_data.hrid AS hrid
, hormone_kits.hormone AS hormone
, hormone_result_data.kit AS kit
, hormone_result_data.assay_date AS assay_date
, hormone_result_data.grams_used AS grams_used
, hormone_result_data.raw_ng_g AS raw_ng_g
, corrected_hormone(hormone_result_data.raw_ng_g, hormone_kits.correction) AS corrected_ng_g
, hormone_result_data.comments AS comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ
JOIN hormone_prep_series
ON hormone_prep_series.tid = hormone_sample_data.tid
JOIN hormone_result_data
ON hormone_result_data.hpsid = hormone_prep_series.hpsid
JOIN hormone_kits
ON hormone_kits.kit = hormone_result_data.kit;
Figure 54. Query Defining the HORMONE_SAMPLES View
SELECT hormone_sample_data.tid AS tid
, hormone_sample_data.hsid AS hsid
, unique_indivs.individ AS individ
, biograph.sname AS sname
, tissue_data.collection_date AS collection_date
, tissue_data.collection_date_status AS collection_date_status
, hormone_sample_data.fzdried_date AS fzdried_date
, hormone_sample_data.sifted_date AS sifted_date
, hormone_sample_data.avail_mass_g AS avail_mass_g
, hormone_sample_data.avail_date AS avail_date
, hormone_sample_data.comments AS comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ;
Figure 56. Query Defining the HUMERUS_STATS View
SELECT humeruses.dartid AS dartid
, count(*) AS husamps
, avg(humeruses.hulength) AS hulength_mean
, stddev(humeruses.hulength) AS hulength_stddev
, avg(humeruses.huunadjusted) AS huunadjusted_mean
, stddev(humeruses.huunadjusted) AS huunadjusted_stddev
FROM humeruses
GROUP BY humeruses.dartid;
Figure 58. Query Defining the INTERACT View
SELECT iid AS iid
, interact_data.sid AS sid
, interact_data.act AS act
, acts.class AS class
, interact_data.date AS date
, julian(interact_data.date) AS jdate
, interact_data.start AS start
, spm(interact_data.start) AS startspm
, stop AS stop
, spm(interact_data.stop) AS stopspm
, interact_data.observer AS observer
, interact_data.handwritten AS handwritten
, interact_data.exact_date AS exact_date
FROM interact_data
JOIN acts
ON (acts.act = interact_data.act);
Figure 60. Query Defining the LOCATIONS_FREE View
SELECT locations.locid AS locid
, locations.institution AS institution
, locations.location AS location
, locations.is_unique AS is_unique
FROM locations
WHERE NOT EXISTS (SELECT 1
FROM tissue_data
WHERE tissue_data.locid = locations.locid)
AND NOT EXISTS (SELECT 1
FROM nucacid_data
WHERE nucacid_data.locid = locations.locid);
Figure 62. Query Defining the MATERNITIES View
SELECT cycles.sname AS mom
, cycles.cid AS cid
, cycles.seq AS seq
, cycles.series AS series
, cycpoints.cpid AS conceive
, cycpoints.date AS zdate
, members.grp AS zdate_grp
, cycpoints.edate AS edate
, cycpoints.ldate AS ldate
, cycpoints.source AS source
, pregs.pid AS pid
, pregs.parity AS parity
, biograph.bioid AS child_bioid
, biograph.sname AS child
, biograph.birth AS birth
FROM cycles
JOIN cycpoints ON (cycpoints.cid = cycles.cid)
JOIN members ON (members.date = cycpoints.date
AND members.sname = cycles.sname)
JOIN pregs ON (pregs.conceive = cycpoints.cpid)
JOIN biograph ON (pregs.pid = biograph.pid);
Figure 64. Query Defining the MIN_MAXS View
SELECT wreadings.wrid AS wrid
, wreadings.wstation AS wstation
, wreadings.wrdaytime AS wrdaytime
, wreadings.estdaytime AS estdaytime
, wreadings.wrperson AS wrperson
, wreadings.wrnotes AS wrnotes
, tempmins.tempmin AS tempmin
, tempmins.curated AS tempmin_curated
, tempmaxs.tempmax AS tempmax
, tempmaxs.curated AS tempmax_curated
, raingauges.rgspan AS rgspan
, raingauges.estrgspan AS estrgspan
, raingauges.rain AS rain
FROM wreadings
LEFT OUTER JOIN tempmins
ON wreadings.wrid = tempmins.wrid
LEFT OUTER JOIN tempmaxs
ON wreadings.wrid = tempmaxs.wrid
LEFT OUTER JOIN raingauges
ON wreadings.wrid = raingauges.wrid;
Figure 66. Query Defining the MIN_MAXS_SORTED View
SELECT wreadings.wrid AS wrid
, wreadings.wstation AS wstation
, wreadings.wrdaytime AS wrdaytime
, wreadings.estdaytime AS estdaytime
, wreadings.wrperson AS wrperson
, wreadings.wrnotes AS wrnotes
, tempmins.tempmin AS tempmin
, tempmins.curated AS tempmin_curated
, tempmaxs.tempmax AS tempmax
, tempmaxs.curated AS tempmax_curated
, raingauges.rgspan AS rgspan
, raingauges.estrgspan AS estrgspan
, raingauges.rain AS rain
FROM wreadings
LEFT OUTER JOIN tempmins
ON wreadings.wrid = tempmins.wrid
LEFT OUTER JOIN tempmaxs
ON wreadings.wrid = tempmaxs.wrid
LEFT OUTER JOIN raingauges
ON wreadings.wrid = raingauges.wrid
ORDER BY wreadings.wrdaytime, wreadings.wstation;;
Figure 68. Query Defining the MPI_EVENTS View
SELECT mpis.mpiid AS mpiid
, mpis.date AS date
, mpis.observer AS observer
, mpis.context_type AS context_type
, mpis.context AS context
, mpi_data.mpidid AS mpidid
, mpi_data.seq AS seq
, mpi_data.mpiact AS mpiact
, actor.mpipid AS actorid
, actor.sname AS actor
, actor.unksname AS unkactor
, actee.mpipid AS acteeid
, actee.sname AS actee
, actee.unksname AS unkactee
, CASE WHEN EXISTS(SELECT 1
FROM mpiacts
WHERE mpiacts.mpiact = mpi_data.mpiact
AND mpiacts.kind = 'H')
THEN
EXISTS(SELECT 1
FROM mpi_data AS request
, mpiacts
, mpi_parts AS requestor
, mpi_parts AS requestee
WHERE request.mpiid = mpi_data.mpiid
AND request.seq < mpi_data.seq
AND mpiacts.mpiact = request.mpiact
AND mpiacts.kind = 'R'
AND requestor.mpidid = request.mpidid
AND requestor.role = 'R'
AND requestor.sname = actee.sname
AND requestee.mpidid = request.mpidid
AND requestee.role = 'E'
AND requestee.sname = actor.sname)
ELSE
NULL
END AS solicited
, EXISTS(SELECT 1
FROM mpi_data AS initial,
mpiacts
WHERE initial.mpiid = mpi_data.mpiid
AND initial.seq = 1
AND mpiacts.mpiact = initial.mpiact
AND mpiacts.decided)
AS decided
, mpi_data.helped AS helped
, mpi_data.active AS active
FROM mpis
LEFT OUTER JOIN mpi_data ON (mpis.mpiid = mpi_data.mpiid)
LEFT OUTER JOIN mpi_parts AS actor
ON (actor.mpidid = mpi_data.mpidid AND actor.role = 'R')
LEFT OUTER JOIN mpi_parts AS actee
ON (actee.mpidid = mpi_data.mpidid AND actee.role = 'E');
Figure 70. Query Defining the MTD_CYCLES View
SELECT cycles.cid AS cid
, cycles.sname AS sname
, cycles.seq AS seq
, cycles.series AS series
, mcp.cpid AS mcpid
, mcp.date AS mdate
, mcp.edate AS emdate
, mcp.ldate AS lmdate
, mcp.source AS msource
, tcp.cpid AS tcpid
, tcp.date AS tdate
, tcp.edate AS etdate
, tcp.ldate AS ltdate
, tcp.source AS tsource
, dcp.cpid AS dcpid
, dcp.date AS ddate
, dcp.edate AS eddate
, dcp.ldate AS lddate
, dcp.source AS dsource
FROM cycles
LEFT OUTER JOIN cycpoints AS mcp ON (mcp.cid = cycles.cid
AND mcp.code = 'M')
LEFT OUTER JOIN cycpoints AS tcp ON (tcp.cid = cycles.cid
AND tcp.code = 'T')
LEFT OUTER JOIN cycpoints AS dcp ON (dcp.cid = cycles.cid
AND dcp.code = 'D')
ORDER BY cycles.sname, cycles.seq;
Figure 72. Query Defining the NUCACID_CONCS View
SELECT nucacid_conc_data.nacid AS nacid
, nucacid_conc_data.naid AS naid
, local_1.localid AS localid_1
, local_2.localid AS localid_2
, nucacid_conc_data.conc_method AS conc_method
, nucacid_conc_methods.descr AS method_descr
, nucacid_conc_data.conc_date AS conc_date
, nucacid_conc_data.quantity AS quantity
, nucacid_conc_data.unit AS unit
FROM nucacid_conc_data
JOIN nucacid_conc_methods
ON nucacid_conc_methods.conc_method = nucacid_conc_data.conc_method
LEFT JOIN nucacid_local_ids AS local_1
ON local_1.naid = nucacid_conc_data.naid
AND local_1.institution = 1
LEFT JOIN nucacid_local_ids AS local_2
ON local_2.naid = nucacid_conc_data.naid
AND local_2.institution = 2;
Figure 74. Query Defining the NUCACIDS View
WITH concat_creators AS (SELECT naid
, string_agg(creator, '/' ORDER BY naid, nacrid) AS created_by
FROM nucacid_creators
GROUP BY naid)
SELECT nucacid_data.naid AS naid
, nucacid_data.tid AS tid
, nucacid_data.locid AS locid
, locations.institution AS institution
, locations.location AS location
, local_1.localid AS localid_1
, local_2.localid AS localid_2
, nucacid_data.uiid AS uiid
, unique_indivs.popid AS popid
, unique_indivs.individ AS individ
, biograph.sname AS sname
, nucacid_data.name_on_tube AS name_on_tube
, nucacid_data.nucacid_type AS nucacid_type
, tissue_data.tissue_type AS tissue_type
, nucacid_data.creation_date AS creation_date
, concat_creators.created_by AS created_by
, nucacid_data.creation_method AS creation_method
, COUNT(nucacid_sources.*) AS na_sources
, nucacid_data.initial_vol_ul AS initial_vol_ul
, nucacid_data.actual_vol_ul AS actual_vol_ul
, nucacid_data.actual_vol_date AS actual_vol_date
, nucacid_data.multi_indivs AS multi_indivs
, nucacid_data.multi_tids AS multi_tids
, nucacid_data.notes AS notes
FROM nucacid_data
JOIN locations
ON locations.locid = nucacid_data.locid
LEFT JOIN tissue_data
ON tissue_data.tid = nucacid_data.tid
LEFT JOIN unique_indivs
ON unique_indivs.uiid = nucacid_data.uiid
LEFT JOIN biograph
ON biograph.bioid::text = unique_indivs.individ
AND unique_indivs.popid = 1
LEFT JOIN nucacid_local_ids AS local_1
ON local_1.naid = nucacid_data.naid
AND local_1.institution = 1
LEFT JOIN nucacid_local_ids AS local_2
ON local_2.naid = nucacid_data.naid
AND local_2.institution = 2
LEFT JOIN nucacid_sources
ON nucacid_sources.naid = nucacid_data.naid
LEFT JOIN concat_creators
ON concat_creators.naid = nucacid_data.naid
GROUP BY nucacid_data.naid
, nucacid_data.tid
, nucacid_data.locid
, locations.institution
, locations.location
, local_1.localid
, local_2.localid
, nucacid_data.uiid
, unique_indivs.popid
, unique_indivs.individ
, biograph.sname
, nucacid_data.name_on_tube
, nucacid_data.nucacid_type
, tissue_data.tissue_type
, nucacid_data.creation_date
, concat_creators.created_by
, nucacid_data.creation_method
, nucacid_data.initial_vol_ul
, nucacid_data.actual_vol_ul
, nucacid_data.actual_vol_date
, nucacid_data.multi_indivs
, nucacid_data.multi_tids
, nucacid_data.notes;
Figure 76. Query Defining the NUCACIDS_W_CONC View
WITH last_quants AS (SELECT DISTINCT
naid
, conc_method
, last_value(quantity) OVER w AS lastquantity
, last_value(unit) OVER w AS lastunit
, last_value(conc_date) OVER w AS lastdate
FROM nucacid_conc_data
WHERE conc_date IS NOT NULL
WINDOW w AS (PARTITION BY naid, conc_method
ORDER BY conc_date
RANGE BETWEEN UNBOUNDED PRECEDING
AND UNBOUNDED FOLLOWING))
, concat_creators AS (SELECT naid
, string_agg(creator, '/' ORDER BY naid, nacrid) AS created_by
FROM nucacid_creators
GROUP BY naid)
SELECT nucacid_data.naid AS naid
, nucacid_data.tid AS tid
, nucacid_data.locid AS locid
, locations.institution AS institution
, locations.location AS location
, local_1.localid AS localid_1
, local_2.localid AS localid_2
, nucacid_data.uiid AS uiid
, unique_indivs.popid AS popid
, unique_indivs.individ AS individ
, biograph.sname AS sname
, nucacid_data.name_on_tube AS name_on_tube
, nucacid_data.nucacid_type AS nucacid_type
, tissue_data.tissue_type AS tissue_type
, nucacid_data.creation_date AS creation_date
, concat_creators.created_by AS created_by
, nucacid_data.creation_method AS creation_method
, COUNT(nucacid_sources.*) AS na_sources
, nucacid_data.initial_vol_ul AS initial_vol_ul
, nucacid_data.actual_vol_ul AS actual_vol_ul
, nucacid_data.actual_vol_date AS actual_vol_date
, nucacid_data.multi_indivs AS multi_indivs
, nucacid_data.multi_tids AS multi_tids
, nucacid_data.notes AS notes
, CASE
WHEN qpcr.lastquantity IS NOT NULL
THEN convert_conc(qpcr.lastquantity, qpcr.lastunit, 'bb_pgul_unit')
ELSE NULL
END AS qpcr_pg_ul
, qpcr.lastdate AS qpcr_lastdate
, CASE
WHEN nanodrop.lastquantity IS NOT NULL
THEN convert_conc(nanodrop.lastquantity, nanodrop.lastunit, 'bb_ngul_unit')
ELSE NULL
END AS nanodrop_ng_ul
, nanodrop.lastdate AS nanodrop_lastdate
, CASE
WHEN qubit.lastquantity IS NOT NULL
THEN convert_conc(qubit.lastquantity, qubit.lastunit, 'bb_ngul_unit')
ELSE NULL
END AS qubit_ng_ul
, qubit.lastdate AS qubit_lastdate
, CASE
WHEN bioanalyzer.lastquantity IS NOT NULL
THEN convert_conc(bioanalyzer.lastquantity, bioanalyzer.lastunit, 'bb_ngul_unit')
ELSE NULL
END AS bioanalyzer_ng_ul
, bioanalyzer.lastdate AS bioanalyzer_lastdate
, CASE
WHEN quantit.lastquantity IS NOT NULL
THEN convert_conc(quantit.lastquantity, quantit.lastunit, 'bb_ngul_unit')
ELSE NULL
END AS quantit_ng_ul
, quantit.lastdate AS quantit_lastdate
FROM nucacid_data
JOIN locations
ON locations.locid = nucacid_data.locid
LEFT JOIN tissue_data
ON tissue_data.tid = nucacid_data.tid
LEFT JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON biograph.bioid::text = unique_indivs.individ
AND unique_indivs.popid = 1
LEFT JOIN nucacid_local_ids AS local_1
ON local_1.naid = nucacid_data.naid
AND local_1.institution = 1
LEFT JOIN nucacid_local_ids AS local_2
ON local_2.naid = nucacid_data.naid
AND local_2.institution = 2
LEFT JOIN nucacid_sources
ON nucacid_sources.naid = nucacid_data.naid
LEFT JOIN concat_creators
ON concat_creators.naid = nucacid_data.naid
LEFT JOIN last_quants AS qpcr
ON qpcr.conc_method = 1
AND qpcr.naid = nucacid_data.naid
LEFT JOIN last_quants AS nanodrop
ON nanodrop.conc_method = 2
AND nanodrop.naid = nucacid_data.naid
LEFT JOIN last_quants AS qubit
ON qubit.conc_method = 3
AND qubit.naid = nucacid_data.naid
LEFT JOIN last_quants AS bioanalyzer
ON bioanalyzer.conc_method = 4
AND bioanalyzer.naid = nucacid_data.naid
LEFT JOIN last_quants AS quantit
ON quantit.conc_method = 5
AND quantit.naid = nucacid_data.naid
GROUP BY nucacid_data.naid
, nucacid_data.tid
, nucacid_data.locid
, locations.institution
, locations.location
, local_1.localid
, local_2.localid
, nucacid_data.uiid
, unique_indivs.popid
, unique_indivs.individ
, biograph.sname
, nucacid_data.name_on_tube
, nucacid_data.nucacid_type
, tissue_data.tissue_type
, nucacid_data.creation_date
, concat_creators.created_by
, nucacid_data.creation_method
, nucacid_data.initial_vol_ul
, nucacid_data.actual_vol_ul
, nucacid_data.actual_vol_date
, nucacid_data.multi_indivs
, nucacid_data.multi_tids
, nucacid_data.notes
, qpcr.lastquantity
, qpcr.lastunit
, qpcr.lastdate
, nanodrop.lastquantity
, nanodrop.lastunit
, nanodrop.lastdate
, qubit.lastquantity
, qubit.lastunit
, qubit.lastdate
, bioanalyzer.lastquantity
, bioanalyzer.lastunit
, bioanalyzer.lastdate
, quantit.lastquantity
, quantit.lastunit
, quantit.lastdate;
Figure 78. Query Defining the PARENTS View
SELECT biograph.sname AS kid
, maternities.mom AS mom
, dads_consensus.dad_consensus AS dad
, maternities.zdate AS zdate
, maternities.zdate_grp AS momgrp
, members.grp AS dadgrp
FROM biograph
LEFT OUTER JOIN maternities
ON (maternities.child = biograph.sname)
LEFT OUTER JOIN dads_consensus
ON (dads_consensus.kid = biograph.sname)
LEFT OUTER JOIN members
ON (members.sname = dads_consensus.dad_consensus
AND members.date = maternities.zdate)
WHERE maternities.mom IS NOT NULL
OR dads_consensus.dad_consensus IS NOT NULL;
Figure 80. Query Defining the PCV_STATS View
SELECT pcvs.dartid AS dartid
, count(*) AS pcvsamps
, avg(pcvs.pcv) AS pcv_mean
, stddev(pcvs.pcv) AS pcv_stddev
FROM pcvs
GROUP BY pcvs.dartid;
Figure 82. Query Defining the POINTS View
SELECT pntid AS pntid
, sid AS sid
, activity AS activity
, posture AS posture
, foodcode AS foodcode
, ptime AS ptime
, spm(ptime) AS ptimespm
FROM point_data;
Figure 84. Query Defining the POTENTIAL_DADS View
SELECT maternities.child_bioid AS bioid
, maternities.child AS kid
, maternities.mom AS mom
, maternities.zdate AS zdate
, maternities.zdate_grp AS grp
, pdads.sname AS pdad
, CASE
WHEN rankdates.ranked <= maternities.zdate
THEN 'A'
WHEN maturedates.matured <= maternities.zdate
THEN 'S'
ELSE 'O'
END
AS status
, maternities.zdate - pdads.birth AS pdad_age_days
, trunc((maternities.zdate - pdads.birth) / 365.25, 1)
AS pdad_age_years
, (SELECT count(*)
FROM members as dadmembers
JOIN members AS mommembers
ON (mommembers.date = dadmembers.date
AND mommembers.supergroup = dadmembers.supergroup)
WHERE dadmembers.sname = pdads.sname
AND dadmembers.date < maternities.zdate
AND dadmembers.date >= maternities.zdate - 5
AND mommembers.sname = maternities.mom
AND mommembers.date < maternities.zdate
AND mommembers.date >= maternities.zdate - 5)
AS estrous_presence
, (SELECT count(*)
FROM actor_actees
WHERE actor_actees.date < maternities.zdate
AND actor_actees.date >= maternities.zdate - 5
AND (actor_actees.act = 'M'
OR actor_actees.act = 'E')
AND actor_actees.actor = pdads.sname
AND actor_actees.actee = maternities.mom)
AS estrous_me
, (SELECT count(*)
FROM actor_actees
WHERE actor_actees.date < maternities.zdate
AND actor_actees.date >= maternities.zdate - 5
AND actor_actees.act = 'C'
AND actor_actees.actor = pdads.sname
AND actor_actees.actee = maternities.mom)
AS estrous_c
FROM maternities
JOIN biograph AS pdads
ON (pdads.sname
IN (SELECT dadmembers.sname
FROM members AS dadmembers
JOIN members AS mommembers
ON (mommembers.date = dadmembers.date
AND mommembers.supergroup
= dadmembers.supergroup)
WHERE dadmembers.sname = pdads.sname
AND dadmembers.date < maternities.zdate
AND dadmembers.date >= maternities.zdate - 5
AND mommembers.sname = maternities.mom
AND mommembers.date < maternities.zdate
AND mommembers.date >= maternities.zdate - 5))
LEFT OUTER JOIN rankdates
ON (rankdates.sname = pdads.sname)
LEFT OUTER JOIN maturedates
ON (maturedates.sname = pdads.sname)
WHERE pdads.sex = 'M'
-- Speed things up by eliminating potential dads
-- who could not possibly interpolate into the mom's group
-- during the fertile period.
AND pdads.statdate >= maternities.zdate - 5 - 14
-- Potential dad must be at least 2192 days old
-- (approximately 6 years) on the zdate.
AND maternities.zdate - pdads.birth >= 2192;
Figure 86. Entity Relationship Diagram of that portion of the POTENTIAL_DADS View which places the mother and potential father in the same group during the fertile period
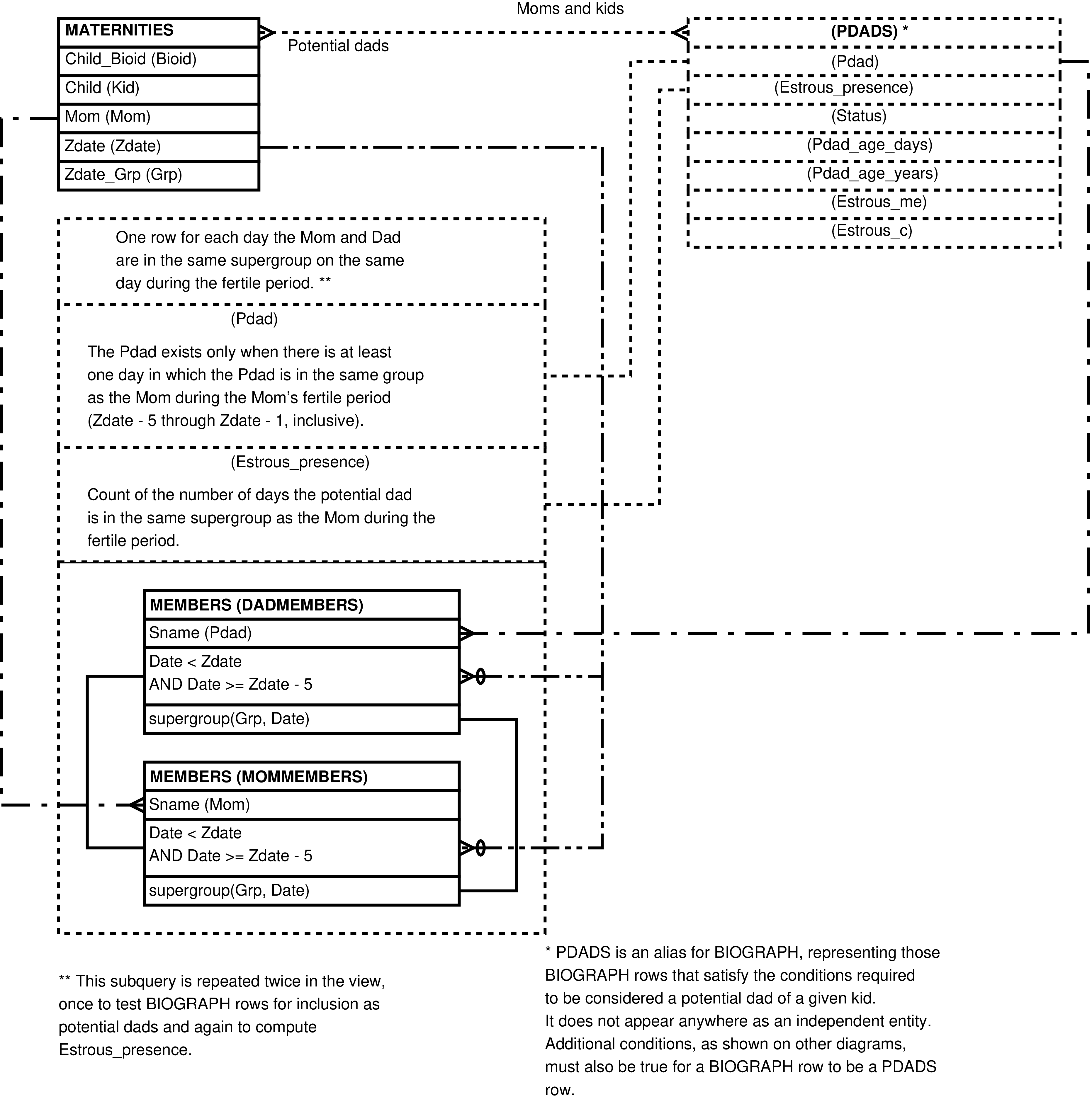 |
Figure 87. Entity Relationship Diagram of that portion of the POTENTIAL_DADS View having easily computed columns
 |
Figure 88. Entity Relationship Diagram of that portion of the POTENTIAL_DADS View involving social interactions
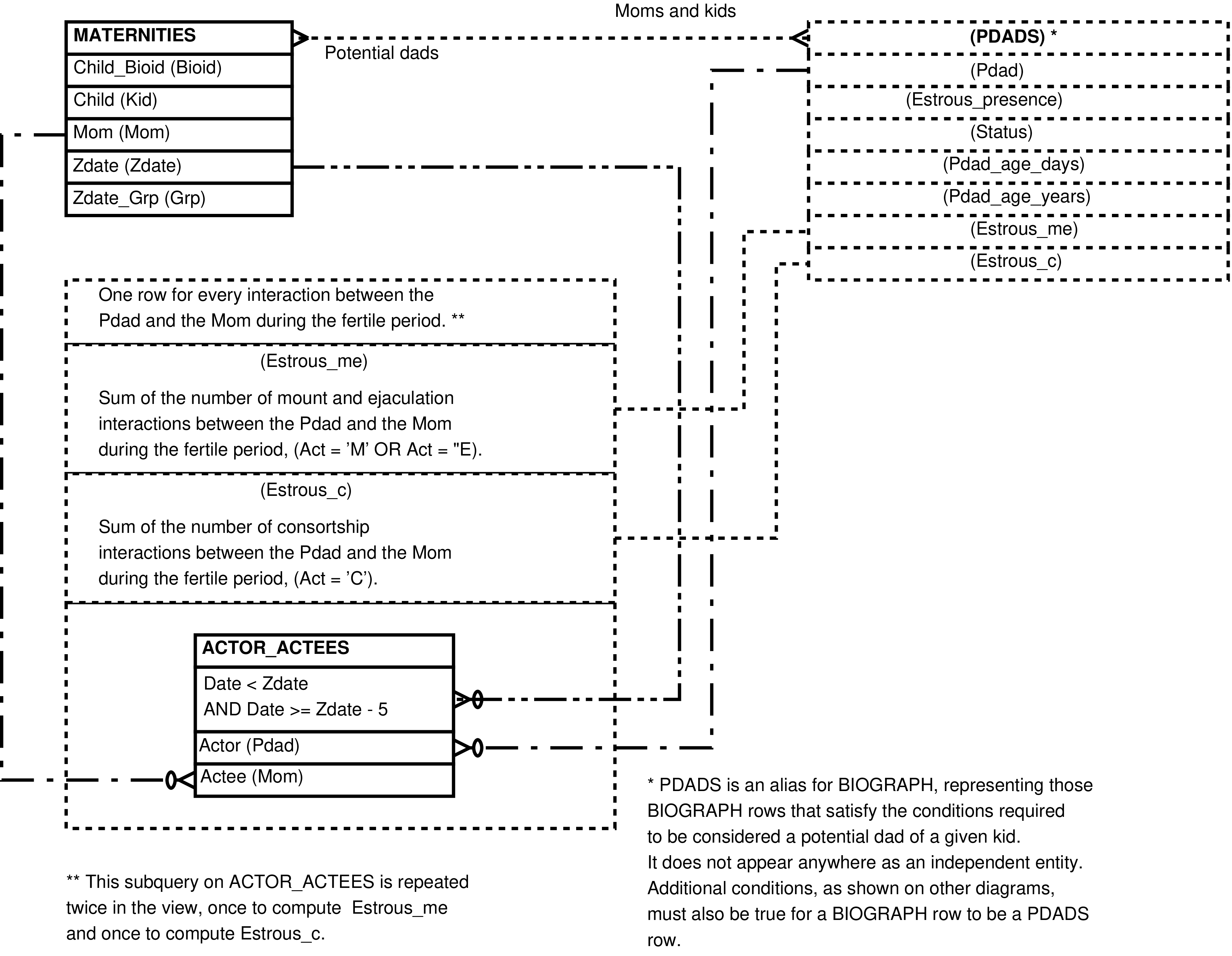 |
Figure 89. Query Defining the PROPORTIONAL_RANKS View
WITH num_indivs AS (
SELECT ranks.rnkdate
, ranks.grp
, ranks.rnktype
, count(*) AS num_members
FROM ranks
GROUP BY ranks.rnkdate, ranks.grp, ranks.rnktype)
SELECT ranks.rnkid AS rnkid
, ranks.sname AS sname
, ranks.rnkdate AS rnkdate
, ranks.grp AS grp
, ranks.rnktype AS rnktype
, ranks.rank AS ordrank
, ranks.ags_density AS ags_density
, ranks.ags_reversals AS ags_reversals
, ranks.ags_expected AS ags_expected
, CASE
WHEN num_indivs.num_members = 1 THEN 1::numeric
ELSE 1 - ((ranks.rank - 1)::numeric / (num_indivs.num_members - 1)::numeric)
END::numeric(5,4) AS proprank
FROM ranks
JOIN num_indivs
ON (num_indivs.rnkdate = ranks.rnkdate
AND num_indivs.grp = ranks.grp
AND num_indivs.rnktype = ranks.rnktype);
Figure 91. Query Defining the QUADS View
SELECT quad_data.quad AS quad
, ST_X(quad_data.xyloc) AS x
, ST_Y(quad_data.xyloc) AS y
, quad_data.aerial AS aerial
FROM quad_data;
Figure 93. Query Defining the SAMPLES_GOFF View
SELECT samples.sid AS sid
, samples.date AS date
, samples.stime AS stime
, samples.observer AS observer
, samples.stype AS stype
, samples.grp AS grp
, samples.sname AS sname
, samples.mins AS mins
, samples.minsis AS minsis
, samples.programid AS programid
, samples.setupid AS setupid
, samples.collection_system AS collection_system
, members.grp AS grp_of_focal
FROM members, samples
WHERE members.sname = samples.sname
AND members.date = CAST(samples.date AS DATE);
Figure 95. Query Defining the SEXSKINS_CYCLES View
SELECT cycles.cid AS cid
, cycles.sname AS sname
, cycles.seq AS seq
, cycles.series AS series
, sexskins.sxid AS sxid
, sexskins.date AS date
, sexskins.size AS size
, sexskins.color AS color
FROM sexskins, cycles
WHERE cycles.cid = sexskins.cid
ORDER BY cycles.sname, sexskins.date;
Figure 97. Query Defining the SEXSKINS_REPRO_NOTES View
SELECT COALESCE(cycles.sname, repro_notes.sname) AS sname
, COALESCE(sexskins.date, repro_notes.date) AS date
, sexskins.cid AS cid
, sexskins.sxid AS sxid
, sexskins.size AS size
, sexskins.color AS color
, repro_notes.rnid AS rnid
, repro_notes.note AS note
FROM sexskins
JOIN cycles
ON cycles.cid = sexskins.cid
FULL OUTER JOIN repro_notes
ON repro_notes.sname = cycles.sname
AND repro_notes.date = sexskins.date;
Figure 99. Query Defining the SWERB View
SELECT swerb_data.swid AS swid
, swerb_departs_data.did AS did
, swerb_departs_data.date AS date
, swerb_data.time AS time
, swerb_bes.beid AS beid
, swerb_bes.focal_grp AS focal_grp
, swerb_bes.seq AS seq
, swerb_data.event AS event
, swerb_data.seen_grp AS seen_grp
, swerb_data.lone_animal AS lone_animal
, swerb_data.quad AS quad
, CASE
WHEN swerb_data.quad IS NOT NULL
THEN 'quad'
WHEN swerb_data.xyloc IS NULL
THEN 'n/a'
ELSE 'gps'
END AS xysource
, COALESCE(ST_X(swerb_data.xyloc), ST_X(quad_data.xyloc))
AS x
, COALESCE(ST_Y(swerb_data.xyloc), ST_Y(quad_data.xyloc))
AS y
, COALESCE(ST_X(ST_TRANSFORM(swerb_data.xyloc, 4326))
, ST_X(ST_TRANSFORM(quad_data.xyloc, 4326)))
AS long
, COALESCE(ST_Y(ST_TRANSFORM(swerb_data.xyloc, 4326))
, ST_Y(ST_TRANSFORM(quad_data.xyloc, 4326)))
AS lat
, swerb_data.altitude AS altitude
, swerb_data.pdop AS pdop
, swerb_data.accuracy AS accuracy
, swerb_data.subgroup AS subgroup
, swerb_data.ogdistance AS ogdistance
, swerb_data.gps_datetime AS gps_datetime
, swerb_data.garmincode AS garmincode
, swerb_data.predator AS predator
, swerb_loc_data.loc AS loc
, swerb_loc_data.adcode AS adcode
, adcodes.adn AS adn
, swerb_loc_data.loc_status AS loc_status
, swerb_loc_data.adtime AS adtime
, ST_X(swerb_loc_gps.xyloc) AS second_x
, ST_Y(swerb_loc_gps.xyloc) AS second_y
, ST_X(ST_TRANSFORM(swerb_loc_gps.xyloc, 4326)) AS second_long
, ST_Y(ST_TRANSFORM(swerb_loc_gps.xyloc, 4326)) AS second_lat
, swerb_loc_gps.altitude AS second_altitude
, swerb_loc_gps.pdop AS second_pdop
, swerb_loc_gps.accuracy AS second_accuracy
, swerb_loc_gps.gps_datetime AS second_gps_datetime
, swerb_loc_gps.garmincode AS second_garmincode
, swerb_bes.start AS start
, swerb_bes.btimeest AS btimeest
, swerb_bes.bsource AS bsource
, swerb_bes.stop AS stop
, swerb_bes.etimeest AS etimeest
, swerb_bes.esource AS esource
, swerb_bes.is_effort AS is_effort
, swerb_departs_gps.gps AS gps
, swerb_bes.notes AS notes
FROM swerb_data
LEFT OUTER JOIN quad_data
ON (quad_data.quad = swerb_data.quad)
JOIN swerb_bes
ON (swerb_bes.beid = swerb_data.beid)
JOIN swerb_departs_data
ON (swerb_departs_data.did = swerb_bes.did)
LEFT OUTER JOIN swerb_departs_gps
ON (swerb_departs_gps.did = swerb_bes.did)
LEFT OUTER JOIN swerb_loc_data
ON (swerb_loc_data.swid = swerb_data.swid)
LEFT OUTER JOIN adcodes ON (adcodes.adcode = swerb_loc_data.adcode)
LEFT OUTER JOIN swerb_loc_gps
ON (swerb_loc_gps.swid = swerb_loc_data.swid);
Figure 101. Query Defining the SWERB_DATA_XY View
SELECT swerb_data.swid AS swid
, swerb_data.beid AS beid
, swerb_data.seen_grp AS seen_grp
, swerb_data.lone_animal AS lone_animal
, swerb_data.event AS event
, swerb_data.time AS time
, swerb_data.quad AS quad
, ST_X(swerb_data.xyloc) AS x
, ST_Y(swerb_data.xyloc) AS y
, ST_X(ST_TRANSFORM(swerb_data.xyloc, 4326)) AS long
, ST_Y(ST_TRANSFORM(swerb_data.xyloc, 4326)) AS lat
, swerb_data.altitude AS altitude
, swerb_data.pdop AS pdop
, swerb_data.accuracy AS accuracy
, swerb_data.subgroup AS subgroup
, swerb_data.ogdistance AS ogdistance
, swerb_data.gps_datetime AS gps_datetime
, swerb_data.garmincode AS garmincode
, swerb_data.predator AS predator
FROM swerb_data;
Figure 103. Query Defining the SWERB_DEPARTS View
SELECT swerb_departs_data.did AS did
, swerb_departs_data.date AS date
, swerb_departs_data.time AS time
, ST_X(swerb_departs_gps.xyloc) AS x
, ST_Y(swerb_departs_gps.xyloc) AS y
, ST_X(ST_TRANSFORM(swerb_departs_gps.xyloc, 4326)) AS long
, ST_Y(ST_TRANSFORM(swerb_departs_gps.xyloc, 4326)) AS lat
, swerb_departs_gps.altitude AS altitude
, swerb_departs_gps.pdop AS pdop
, swerb_departs_gps.accuracy AS accuracy
, swerb_departs_gps.gps AS gps
, swerb_departs_gps.garmincode AS garmincode
FROM swerb_departs_data
LEFT OUTER JOIN swerb_departs_gps
ON (swerb_departs_gps.did = swerb_departs_data.did);
Figure 105. Query Defining the SWERB_GW_LOC_DATA_XY View
SELECT swerb_gw_loc_data.sgwlid AS sgwlid
, swerb_gw_loc_data.loc AS loc
, swerb_gw_loc_data.date AS date
, swerb_gw_loc_data.time AS time
, swerb_gw_loc_data.quad AS quad
, swerb_gw_loc_data.xysource AS xysource
, ST_X(swerb_gw_loc_data.xyloc) AS x
, ST_Y(swerb_gw_loc_data.xyloc) AS y
, ST_X(ST_TRANSFORM(swerb_gw_loc_data.xyloc, 4326)) AS long
, ST_Y(ST_TRANSFORM(swerb_gw_loc_data.xyloc, 4326)) AS lat
, swerb_gw_loc_data.altitude AS altitude
, swerb_gw_loc_data.pdop AS pdop
, swerb_gw_loc_data.accuracy AS accuracy
, swerb_gw_loc_data.gps AS gps
, swerb_gw_loc_data.notes AS notes
FROM swerb_gw_loc_data;
Figure 107. Query Defining the SWERB_GW_LOCS View
SELECT swerb_gw_loc_data.sgwlid AS sgwlid
, swerb_gw_loc_data.loc AS loc
, swerb_gw_loc_data.date AS date
, swerb_gw_loc_data.time AS time
, swerb_gw_loc_data.quad AS quad
, CASE
WHEN swerb_gw_loc_data.xyloc IS NULL
THEN 'quad'
ELSE swerb_gw_loc_data.xysource
END AS xysource
, COALESCE(ST_X(swerb_gw_loc_data.xyloc), ST_X(quad_data.xyloc))
AS x
, COALESCE(ST_Y(swerb_gw_loc_data.xyloc), ST_Y(quad_data.xyloc))
AS y
, COALESCE(ST_X(ST_TRANSFORM(swerb_gw_loc_data.xyloc, 4326))
, ST_X(ST_TRANSFORM(quad_data.xyloc, 4326)))
AS long
, COALESCE(ST_Y(ST_TRANSFORM(swerb_gw_loc_data.xyloc, 4326))
, ST_Y(ST_TRANSFORM(quad_data.xyloc, 4326)))
AS lat
, swerb_gw_loc_data.altitude AS altitude
, swerb_gw_loc_data.pdop AS pdop
, swerb_gw_loc_data.accuracy AS accuracy
, swerb_gw_loc_data.gps AS gps
, swerb_gw_loc_data.notes AS notes
FROM swerb_gw_loc_data
LEFT OUTER JOIN quad_data
ON (quad_data.quad = swerb_gw_loc_data.quad);
Figure 109. Query Defining the SWERB_LOC_GPS_XY View
SELECT swerb_loc_gps.swid AS swid
, ST_X(swerb_loc_gps.xyloc) AS x
, ST_Y(swerb_loc_gps.xyloc) AS y
, ST_X(ST_TRANSFORM(swerb_loc_gps.xyloc, 4326)) AS long
, ST_Y(ST_TRANSFORM(swerb_loc_gps.xyloc, 4326)) AS lat
, swerb_loc_gps.altitude AS altitude
, swerb_loc_gps.pdop AS pdop
, swerb_loc_gps.accuracy AS accuracy
, swerb_loc_gps.gps_datetime AS gps_datetime
, swerb_loc_gps.garmincode AS garmincode
FROM swerb_loc_gps;
Figure 111. Query Defining the SWERB_LOCS View
SELECT swerb_loc_data.swid AS swid
, swerb_loc_data.loc AS loc
, swerb_loc_data.adcode AS adcode
, adcodes.adn AS adn
, swerb_loc_data.loc_status AS loc_status
, swerb_loc_data.adtime AS time
FROM swerb_loc_data
JOIN adcodes ON (adcodes.adcode = swerb_loc_data.adcode);
Figure 113. Query Defining the SWERB_UPLOAD View
SELECT NULL::TEXT AS header
, NULL::TEXT AS name
, NULL::TEXT AS description
, NULL::TEXT AS type
, NULL::TEXT AS position
, NULL::TEXT AS altitude
, NULL::TEXT AS depth
, NULL::TEXT AS proximity
, NULL::TEXT AS display_mode
, NULL::TEXT AS color
, NULL::TEXT AS symbol
, NULL::TEXT AS facility
, NULL::TEXT AS city
, NULL::TEXT AS state
, NULL::TEXT AS country
, NULL::TEXT AS pdop
, NULL::TEXT AS accuracy
, NULL::TEXT AS quad
, NULL::TEXT AS date
, NULL::TEXT AS timeest
, NULL::TEXT AS source
, NULL::TEXT AS lone_animal
, NULL::TEXT AS is_effort
, NULL::BOOLEAN AS secondary_ad
, NULL::BOOLEAN AS be_has_coords
, NULL::TEXT AS notes
WHERE _raise_babase_exception(
'Cannot select SWERB_UPLOAD'
|| ': The only use of the SWERB_UPLOAD view is to insert'
|| ' new data into the SWERB portion of babase');
Figure 114. Entity Relationship Diagram of the SWERB_UPLOAD View
Figure 115. Query Defining the TESTES_ARC_STATS View
SELECT testesdartids.dartid AS dartid
, testesllength.testllengthsamps AS testllengthsamps
, testesllength.testllength_mean AS testllength_mean
, testesllength.testllength_stddev AS testllength_stddev
, testeslwidth.testlwidthsamps AS testlwidthsamps
, testeslwidth.testlwidth_mean AS testlwidth_mean
, testeslwidth.testlwidth_stddev AS testlwidth_stddev
, testesrlength.testrlengthsamps AS testrlengthsamps
, testesrlength.testrlength_mean AS testrlength_mean
, testesrlength.testrlength_stddev AS testrlength_stddev
, testesrwidth.testrwidthsamps AS testrwidthsamps
, testesrwidth.testrwidth_mean AS testrwidth_mean
, testesrwidth.testrwidth_stddev AS testrwidth_stddev
FROM (SELECT testes_arc.dartid
FROM testes_arc
GROUP BY testes_arc.dartid)
AS testesdartids
LEFT OUTER JOIN
(SELECT testes_arc.dartid AS llengthdartid
, count(*) AS testllengthsamps
, avg(testes_arc.testlength) AS testllength_mean
, stddev(testes_arc.testlength) AS testllength_stddev
FROM testes_arc
WHERE testes_arc.testside = 'L'
AND testes_arc.testlength IS NOT NULL
GROUP BY testes_arc.dartid)
AS testesllength
ON testesllength.llengthdartid = testesdartids.dartid
LEFT OUTER JOIN
(SELECT testes_arc.dartid AS lwidthdartid
, count(*) AS testlwidthsamps
, avg(testes_arc.testwidth) AS testlwidth_mean
, stddev(testes_arc.testwidth) AS testlwidth_stddev
FROM testes_arc
WHERE testes_arc.testside = 'L'
AND testes_arc.testwidth IS NOT NULL
GROUP BY testes_arc.dartid)
AS testeslwidth
ON testeslwidth.lwidthdartid = testesdartids.dartid
LEFT OUTER JOIN
(SELECT testes_arc.dartid AS rlengthdartid
, count(*) AS testrlengthsamps
, avg(testes_arc.testlength) AS testrlength_mean
, stddev(testes_arc.testlength) AS testrlength_stddev
FROM testes_arc
WHERE testes_arc.testside = 'R'
AND testes_arc.testlength IS NOT NULL
GROUP BY testes_arc.dartid)
AS testesrlength
ON testesrlength.rlengthdartid = testesdartids.dartid
LEFT OUTER JOIN
(SELECT testes_arc.dartid AS rwidthdartid
, count(*) AS testrwidthsamps
, avg(testes_arc.testwidth) AS testrwidth_mean
, stddev(testes_arc.testwidth) AS testrwidth_stddev
FROM testes_arc
WHERE testes_arc.testside = 'R'
AND testes_arc.testwidth IS NOT NULL
GROUP BY testes_arc.dartid)
AS testesrwidth
ON testesrwidth.rwidthdartid = testesdartids.dartid;
Figure 117. Query Defining the TESTES_DIAM_STATS View
SELECT testesdartids.dartid AS dartid
, testesllength.testllengthsamps AS testllengthsamps
, testesllength.testllength_mean AS testllength_mean
, testesllength.testllength_stddev AS testllength_stddev
, testeslwidth.testlwidthsamps AS testlwidthsamps
, testeslwidth.testlwidth_mean AS testlwidth_mean
, testeslwidth.testlwidth_stddev AS testlwidth_stddev
, testesrlength.testrlengthsamps AS testrlengthsamps
, testesrlength.testrlength_mean AS testrlength_mean
, testesrlength.testrlength_stddev AS testrlength_stddev
, testesrwidth.testrwidthsamps AS testrwidthsamps
, testesrwidth.testrwidth_mean AS testrwidth_mean
, testesrwidth.testrwidth_stddev AS testrwidth_stddev
FROM (SELECT testes_diam.dartid
FROM testes_diam
GROUP BY testes_diam.dartid)
AS testesdartids
LEFT OUTER JOIN
(SELECT testes_diam.dartid AS llengthdartid
, count(*) AS testllengthsamps
, avg(testes_diam.testlength) AS testllength_mean
, stddev(testes_diam.testlength) AS testllength_stddev
FROM testes_diam
WHERE testes_diam.testside = 'L'
AND testes_diam.testlength IS NOT NULL
GROUP BY testes_diam.dartid)
AS testesllength
ON testesllength.llengthdartid = testesdartids.dartid
LEFT OUTER JOIN
(SELECT testes_diam.dartid AS lwidthdartid
, count(*) AS testlwidthsamps
, avg(testes_diam.testwidth) AS testlwidth_mean
, stddev(testes_diam.testwidth) AS testlwidth_stddev
FROM testes_diam
WHERE testes_diam.testside = 'L'
AND testes_diam.testwidth IS NOT NULL
GROUP BY testes_diam.dartid)
AS testeslwidth
ON testeslwidth.lwidthdartid = testesdartids.dartid
LEFT OUTER JOIN
(SELECT testes_diam.dartid AS rlengthdartid
, count(*) AS testrlengthsamps
, avg(testes_diam.testlength) AS testrlength_mean
, stddev(testes_diam.testlength) AS testrlength_stddev
FROM testes_diam
WHERE testes_diam.testside = 'R'
AND testes_diam.testlength IS NOT NULL
GROUP BY testes_diam.dartid)
AS testesrlength
ON testesrlength.rlengthdartid = testesdartids.dartid
LEFT OUTER JOIN
(SELECT testes_diam.dartid AS rwidthdartid
, count(*) AS testrwidthsamps
, avg(testes_diam.testwidth) AS testrwidth_mean
, stddev(testes_diam.testwidth) AS testrwidth_stddev
FROM testes_diam
WHERE testes_diam.testside = 'R'
AND testes_diam.testwidth IS NOT NULL
GROUP BY testes_diam.dartid)
AS testesrwidth
ON testesrwidth.rwidthdartid = testesdartids.dartid;
Figure 119. Query Defining the TESTOSTERONES View
SELECT hormone_sample_data.tid
, hormone_prep_series.hpsid
, hormone_result_data.hrid
, hormone_sample_data.hsid
, biograph.sname
, tissue_data.collection_date
, tissue_data.collection_date_status AS collection_date_status
, hormone_sample_data.fzdried_date AS fzdried_date
, hormone_sample_data.sifted_date AS sifted_date
, meoh_ext.procedure_date AS me_extracted
, spe.procedure_date AS sp_extracted
, hormone_result_data.raw_ng_g AS raw_ng_g
, corrected_hormone(hormone_result_data.raw_ng_g, hormone_kits.correction) AS corrected_ng_g
, hormone_result_data.assay_date
, hormone_kits.hormone AS hormone
, hormone_result_data.kit AS kit
, hormone_sample_data.comments AS sample_comments
, hormone_result_data.comments AS result_comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ
JOIN hormone_prep_series
ON hormone_prep_series.tid = hormone_sample_data.tid
JOIN hormone_result_data
ON hormone_result_data.hpsid = hormone_prep_series.hpsid
JOIN hormone_kits
ON hormone_kits.kit = hormone_result_data.kit
AND hormone_kits.correction IS NOT NULL
AND hormone_kits.hormone = 'T'
LEFT JOIN hormone_prep_data AS meoh_ext
ON meoh_ext.procedure = 'MEOH_EXT'
AND meoh_ext.hpsid = hormone_prep_series.hpsid
LEFT JOIN hormone_prep_data AS spe
ON spe.procedure = 'SPE'
AND spe.hpsid = hormone_prep_series.hpsid;
Figure 121. Query Defining the THYROID_HORMONES View
SELECT hormone_sample_data.tid
, hormone_prep_series.hpsid
, hormone_result_data.hrid
, hormone_sample_data.hsid
, biograph.sname
, tissue_data.collection_date
, tissue_data.collection_date_status AS collection_date_status
, hormone_sample_data.fzdried_date AS fzdried_date
, hormone_sample_data.sifted_date AS sifted_date
, etoh_ext.procedure_date AS et_extracted
, hormone_result_data.raw_ng_g AS raw_ng_g
, corrected_hormone(hormone_result_data.raw_ng_g, hormone_kits.correction) AS corrected_ng_g
, hormone_result_data.assay_date
, hormone_kits.hormone AS hormone
, hormone_result_data.kit AS kit
, hormone_sample_data.comments AS sample_comments
, hormone_result_data.comments AS result_comments
FROM hormone_sample_data
JOIN tissue_data
ON tissue_data.tid = hormone_sample_data.tid
JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON unique_indivs.popid = 1
AND biograph.bioid::text = unique_indivs.individ
JOIN hormone_prep_series
ON hormone_prep_series.tid = hormone_sample_data.tid
JOIN hormone_result_data
ON hormone_result_data.hpsid = hormone_prep_series.hpsid
JOIN hormone_kits
ON hormone_kits.kit = hormone_result_data.kit
AND hormone_kits.correction IS NOT NULL
AND hormone_kits.hormone = 'TH'
LEFT JOIN hormone_prep_data AS etoh_ext
ON etoh_ext.procedure = 'ETOH_EXT'
AND etoh_ext.hpsid = hormone_prep_series.hpsid;
Figure 123. Query Defining the TISSUES View
SELECT tissue_data.tid AS tid
, tissue_data.locid AS locid
, locations.institution AS institution
, locations.location AS location
, local_1.localid AS localid_1
, local_2.localid AS localid_2
, tissue_data.uiid AS uiid
, unique_indivs.popid AS popid
, unique_indivs.individ AS individ
, biograph.sname AS sname
, tissue_data.name_on_tube AS name_on_tube
, tissue_data.collection_date AS collection_date
, tissue_data.collection_time AS collection_time
, tissue_data.tissue_type AS tissue_type
, tissue_data.storage_medium AS storage_medium
, tissue_data.misid_status AS misid_status
, tissue_data.collection_date_status AS collection_date_status
, tissue_data.multi_indivs AS multi_indivs
, COUNT(tissue_sources.*) AS tissue_sources
, tissue_data.notes AS notes
FROM tissue_data
JOIN locations
ON locations.locid = tissue_data.locid
LEFT JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON biograph.bioid::text = unique_indivs.individ
AND unique_indivs.popid = 1
LEFT JOIN tissue_local_ids AS local_1
ON local_1.tid = tissue_data.tid
AND local_1.institution = 1
LEFT JOIN tissue_local_ids AS local_2
ON local_2.tid = tissue_data.tid
AND local_2.institution = 2
LEFT JOIN tissue_sources
ON tissue_sources.tid = tissue_data.tid
GROUP BY tissue_data.tid
, tissue_data.locid
, locations.institution
, locations.location
, local_1.localid
, local_2.localid
, tissue_data.uiid
, unique_indivs.popid
, unique_indivs.individ
, biograph.sname
, tissue_data.name_on_tube
, tissue_data.collection_date
, tissue_data.collection_time
, tissue_data.tissue_type
, tissue_data.storage_medium
, tissue_data.misid_status
, tissue_data.collection_date_status
, tissue_data.multi_indivs
, tissue_data.notes;
Figure 125. Query Defining the TISSUES_HORMONES View
SELECT tissue_data.tid AS tid
, tissue_data.locid AS locid
, locations.institution AS institution
, locations.location AS location
, local_1.localid AS localid_1
, local_2.localid AS localid_2
, tissue_data.uiid AS uiid
, unique_indivs.popid AS popid
, unique_indivs.individ AS individ
, biograph.sname AS sname
, tissue_data.name_on_tube AS name_on_tube
, tissue_data.collection_date AS collection_date
, tissue_data.collection_time AS collection_time
, tissue_data.tissue_type AS tissue_type
, tissue_data.storage_medium AS storage_medium
, tissue_data.misid_status AS misid_status
, tissue_data.collection_date_status AS collection_date_status
, tissue_data.multi_indivs AS multi_indivs
, COUNT(tissue_sources.*) AS tissue_sources
, tissue_data.notes AS notes
, hormone_sample_data.hsid AS hsid
, hormone_sample_data.fzdried_date AS fzdried_date
, hormone_sample_data.sifted_date AS sifted_date
, hormone_sample_data.avail_mass_g AS avail_mass_g
, hormone_sample_data.avail_date AS avail_date
, hormone_sample_data.comments AS comments
FROM tissue_data
JOIN locations
ON locations.locid = tissue_data.locid
LEFT JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON biograph.bioid::text = unique_indivs.individ
AND unique_indivs.popid = 1
LEFT JOIN tissue_local_ids AS local_1
ON local_1.tid = tissue_data.tid
AND local_1.institution = 1
LEFT JOIN tissue_local_ids AS local_2
ON local_2.tid = tissue_data.tid
AND local_2.institution = 2
LEFT JOIN tissue_sources
ON tissue_sources.tid = tissue_data.tid
LEFT JOIN hormone_sample_data
ON hormone_sample_data.tid = tissue_data.tid
GROUP BY tissue_data.tid
, tissue_data.locid
, locations.institution
, locations.location
, local_1.localid
, local_2.localid
, tissue_data.uiid
, unique_indivs.popid
, unique_indivs.individ
, biograph.sname
, tissue_data.name_on_tube
, tissue_data.collection_date
, tissue_data.collection_time
, tissue_data.tissue_type
, tissue_data.storage_medium
, tissue_data.misid_status
, tissue_data.collection_date_status
, tissue_data.multi_indivs
, tissue_data.notes
, hormone_sample_data.hsid
, hormone_sample_data.fzdried_date
, hormone_sample_data.sifted_date
, hormone_sample_data.avail_mass_g
, hormone_sample_data.avail_date
, hormone_sample_data.comments;
Figure 127. Query Defining the ULNA_STATS View
SELECT ulnas.dartid AS dartid
, count(*) AS ulsamps
, avg(ulnas.ullength) AS ullength_mean
, stddev(ulnas.ullength) AS ullength_stddev
, avg(ulnas.ulunadjusted) AS ulunadjusted_mean
, stddev(ulnas.ulunadjusted) AS ulunadjusted_stddev
FROM ulnas
GROUP BY ulnas.dartid;
Figure 129. Query Defining the VAGINAL_PH_STATS View
SELECT vaginal_phs.dartid AS dartid
, count(*) AS vpsamps
, avg(vaginal_phs.ph) AS vp_mean
, stddev(vaginal_phs.ph) AS vp_stddev
FROM vaginal_phs
GROUP BY vaginal_phs.dartid;
Figure 131. Query Defining the WOUNDSPATHOLOGIES View
WITH concat_observers AS (SELECT wprid
, string_agg(observer, '/' ORDER BY wpoid) as observers
FROM wp_observers
GROUP BY wprid)
SELECT wp_reports.wprid AS wprid
, wp_reports.wid AS wid
, wp_reports.date AS reportdate
, wp_reports.time AS reporttime
, concat_observers.observers AS observers
, wp_reports.sname AS sname
, wp_reports.grp AS grp
, wp_reports.observercomments AS observercomments
, wp_reports.reportstate AS reportstate
, wp_details.wpdid AS wpdid
, wp_details.woundpathcode AS woundpathcode
, wp_details.cluster AS cluster
, wp_details.maxdimension AS maxdimension
, wp_details.impairslocomotion AS impairslocomotion
, wp_details.infectionsigns AS infectionsigns
, wp_details.notes AS detailnotes
, wp_affectedparts.wpaid AS wpaid
, wp_affectedparts.bodypart AS bodypart
, bodyparts.bodyside AS bodyside
, bodyparts.innerouter AS innerouter
, bodyparts.bodyregion AS bodyregion
, wp_affectedparts.quantity_affecting_part AS quantity_affecting_part
FROM wp_reports
LEFT JOIN concat_observers
ON concat_observers.wprid = wp_reports.wprid
LEFT JOIN wp_details
ON wp_details.wprid = wp_reports.wprid
LEFT JOIN wp_affectedparts
ON wp_affectedparts.wpdid = wp_details.wpdid
LEFT JOIN bodyparts
ON bodyparts.bpid = wp_affectedparts.bodypart;
Figure 133. Query Defining the WP_DETAILS_AFFECTEDPARTS View
SELECT wp_details.wpdid AS wpdid
, wp_reports.wprid AS wprid
, wp_reports.wid AS wid
, wp_details.woundpathcode AS woundpathcode
, wp_details.cluster AS cluster
, wp_details.maxdimension AS maxdimension
, wp_details.impairslocomotion AS impairslocomotion
, wp_details.infectionsigns AS infectionsigns
, wp_details.notes AS detailnotes
, wp_affectedparts.wpaid AS wpaid
, wp_affectedparts.wpdid AS bodypart_wpdid
, wp_affectedparts.bodypart AS bodypart
, bodyparts.bodyside AS bodyside
, bodyparts.innerouter AS innerouter
, bodyparts.bodyregion AS bodyregion
, wp_affectedparts.quantity_affecting_part AS quantity_affecting_part
FROM wp_reports
JOIN wp_details
ON wp_details.wprid = wp_reports.wprid
LEFT JOIN wp_affectedparts
ON wp_affectedparts.wpdid = wp_details.wpdid
LEFT JOIN bodyparts
ON bodyparts.bpid = wp_affectedparts.bodypart;
Figure 135. Query Defining the WP_HEALS View
WITH concat_observers AS (SELECT wprid
, string_agg(observer, '/' ORDER BY wpoid) as observers
FROM wp_observers
GROUP BY wprid)
SELECT wp_reports.wprid AS wprid
, wp_reports.wid AS wid
, wp_reports.date AS reportdate
, wp_reports.time AS reporttime
, concat_observers.observers AS observers
, wp_reports.sname AS sname
, wp_reports.grp AS grp
, wp_reports.observercomments AS observercomments
, wp_reports.reportstate AS reportstate
, wp_details.wpdid AS wpdid
, wp_details.woundpathcode AS woundpathcode
, wp_details.cluster AS cluster
, wp_details.maxdimension AS maxdimension
, wp_details.impairslocomotion AS impairslocomotion
, wp_details.infectionsigns AS infectionsigns
, wp_details.notes AS detailnotes
, wp_affectedparts.wpaid AS wpaid
, wp_affectedparts.bodypart AS bodypart
, bodyparts.bodyside AS bodyside
, bodyparts.innerouter AS innerouter
, bodyparts.bodyregion AS bodyregion
, wp_affectedparts.quantity_affecting_part AS quantity_affecting_part
, wp_healupdates.wphid AS wphid
, wp_healupdates.date AS healdate
, wp_healupdates.healstatus AS healstatus
, wp_healupdates.notes AS healnotes
FROM wp_healupdates
LEFT JOIN wp_affectedparts
ON wp_affectedparts.wpaid = wp_healupdates.wpaid
LEFT JOIN bodyparts
ON bodyparts.bpid = wp_affectedparts.bodypart
LEFT JOIN wp_details
ON wp_details.wpdid = COALESCE(wp_affectedparts.wpdid, wp_healupdates.wpdid)
LEFT JOIN wp_reports
ON wp_reports.wprid = COALESCE(wp_details.wprid, wp_healupdates.wprid)
LEFT JOIN concat_observers
ON concat_observers.wprid = wp_reports.wprid;
Figure 137. Entity Relationship Diagram of the WP_HEALS View for rows with an update to a wound/pathology report
 |
Figure 138. Entity Relationship Diagram of the WP_HEALS View for rows with an update to a wound/pathology cluster
 |
Figure 139. Entity Relationship Diagram of the WP_HEALS View for rows with an update to an affected body part
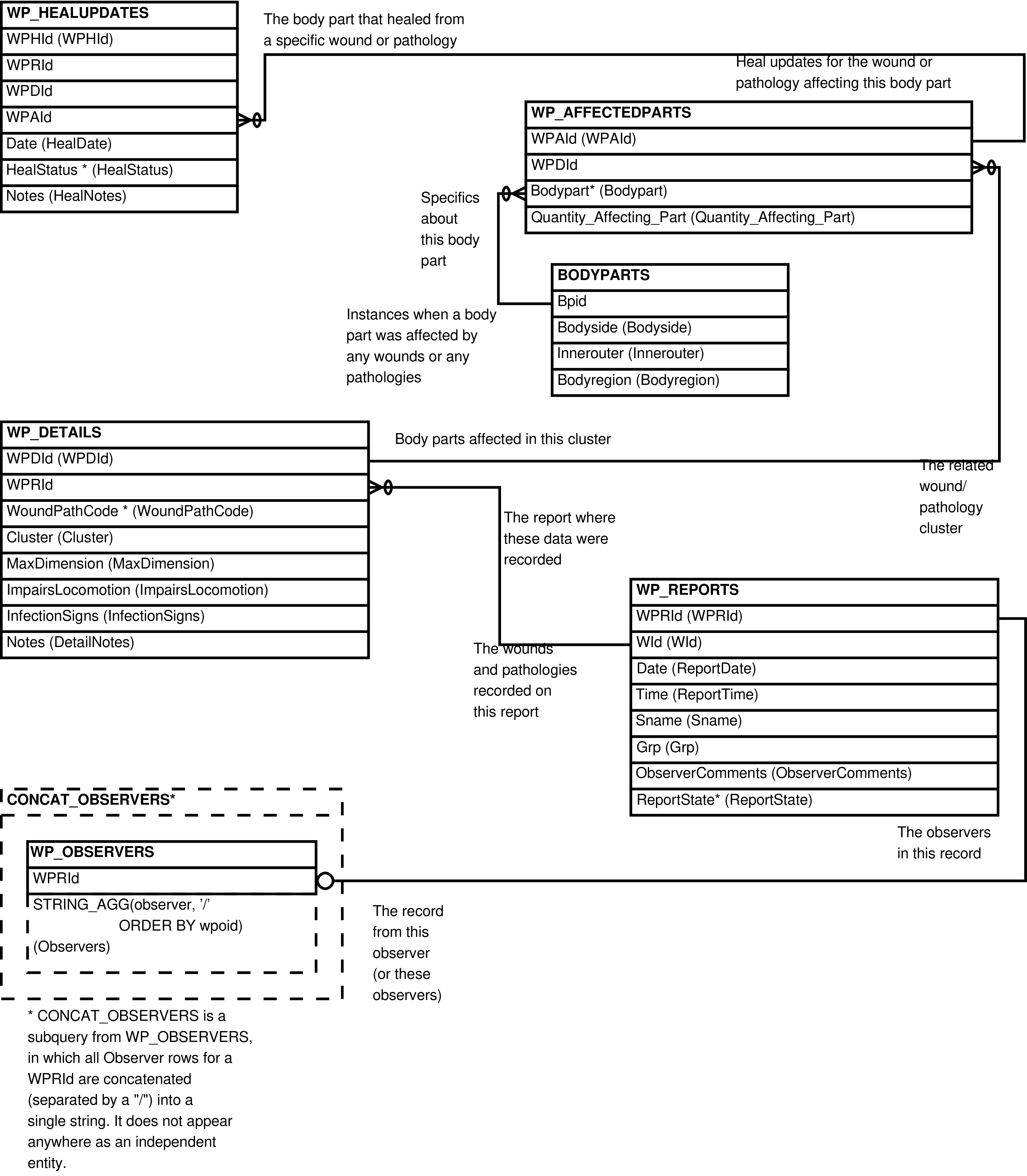 |
Figure 140. Query Defining the WP_REPORTS_OBSERVERS View
WITH concat_observers AS (SELECT wprid
, string_agg(observer, '/' ORDER BY wpoid) as observers
FROM wp_observers
GROUP BY wprid)
SELECT wp_reports.wprid AS wprid
, wp_reports.wid AS wid
, wp_reports.date AS date
, wp_reports.time AS time
, concat_observers.observers AS observers
, wp_reports.sname AS sname
, wp_reports.grp AS grp
, wp_reports.observercomments AS observercomments
, wp_reports.reportstate AS reportstate
FROM wp_reports
LEFT JOIN concat_observers
ON concat_observers.wprid = wp_reports.wprid;
In addition to the above views there are a number of views
which produce the group of a referenced individual as of a
pertinent date. These views are all named after the table from
which they are derived, with the addition of the suffixed
_GRP. They are nearly identical to the table
from which they derive, differing only by the addition of a
column named Grp.
The only operation allowed on these views is SELECT. INSERT, UPDATE, and DELETE are not allowed.
Figure 142. Query Defining the BIRTH_GRP View
SELECT biograph.*
, members.grp AS grp
FROM members, biograph
WHERE members.sname = biograph.sname
AND members.date = CAST(biograph.birth AS DATE);
Figure 144. Query Defining the ENTRYDATE_GRP View
SELECT biograph.*
, members.grp AS grp
FROM members, biograph
WHERE members.sname = biograph.sname
AND members.date = CAST(biograph.entrydate AS DATE);
Figure 146. Query Defining the STATDATE_GRP View
SELECT biograph.*
, members.grp AS grp
FROM members, biograph
WHERE members.sname = biograph.sname
AND members.date = CAST(biograph.statdate AS DATE);
Figure 148. Query Defining the CONSORTDATES_GRP View
SELECT consortdates.*
, members.grp AS grp
FROM members, consortdates
WHERE members.sname = consortdates.sname
AND members.date = CAST(consortdates.consorted AS DATE);
Figure 150. Query Defining the CYCGAPDAYS_GRP View
SELECT cycgapdays.*
, members.grp AS grp
FROM members, cycgapdays
WHERE members.sname = cycgapdays.sname
AND members.date = CAST(cycgapdays.date AS DATE);
Figure 152. Query Defining the CYCGAPS_GRP View
SELECT cycgaps.*
, members.grp AS grp
FROM members, cycgaps
WHERE members.sname = cycgaps.sname
AND members.date = CAST(cycgaps.date AS DATE);
Figure 154. Query Defining the CYCSTATS_GRP View
SELECT cycstats.*
, members.grp AS grp
FROM members, cycstats
WHERE members.sname = cycstats.sname
AND members.date = CAST(cycstats.date AS DATE);
Figure 156. Query Defining the DARTINGS_GRP View
SELECT dartings.*
, members.grp AS grp
FROM members, dartings
WHERE members.sname = dartings.sname
AND members.date = CAST(dartings.date AS DATE);
Figure 158. Query Defining the DISPERSEDATES_GRP View
SELECT dispersedates.*
, members.grp AS grp
FROM members, dispersedates
WHERE members.sname = dispersedates.sname
AND members.date = CAST(dispersedates.dispersed AS DATE);
Figure 160. Query Defining the MATUREDATES_GRP View
SELECT maturedates.*
, members.grp AS grp
FROM members, maturedates
WHERE members.sname = maturedates.sname
AND members.date = CAST(maturedates.matured AS DATE);
Figure 162. Query Defining the MDINTERVALS_GRP View
SELECT mdintervals.*
, members.grp AS grp
FROM members, mdintervals
WHERE members.sname = mdintervals.sname
AND members.date = CAST(mdintervals.date AS DATE);
Figure 164. Query Defining the MMINTERVALS_GRP View
SELECT mmintervals.*
, members.grp AS grp
FROM members, mmintervals
WHERE members.sname = mmintervals.sname
AND members.date = CAST(mmintervals.date AS DATE);