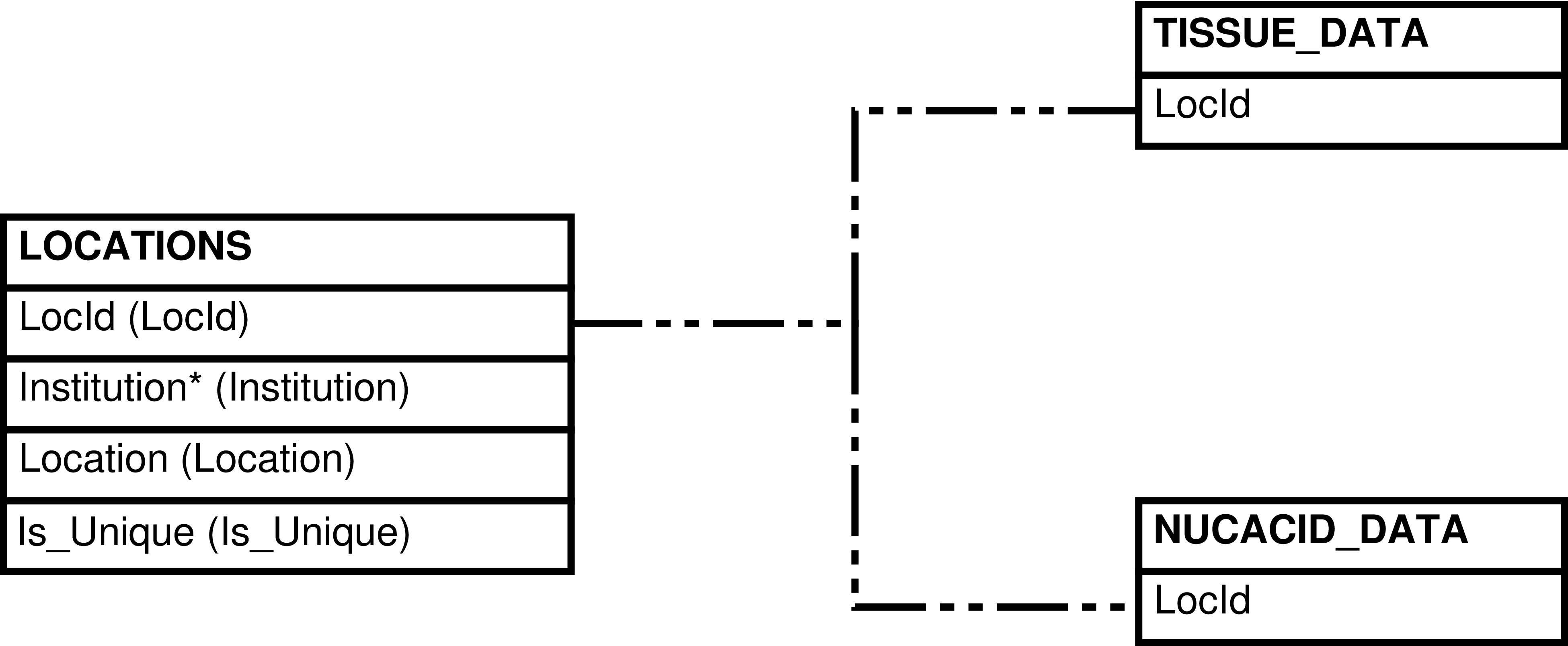
Contains one row for every LOCATIONS row whose Location is not used in NUCACID_DATA or TISSUE_DATA. That is, it contains one row for every location that is not occupied by a nucleic acid or tissue sample.
Tip
Use this view when looking for locations available to store new samples.
Caution
This view makes no attempt to treat non-unique
locations (those whose Is_Unique
is FALSE) differently from unique ones. A non-unique
location that might in reality be available will not appear
in this view if it is already in use in NUCACID_DATA or TISSUE_DATA.
Figure 6.130. Query Defining the LOCATIONS_FREE View
SELECT locations.locid AS locid
, locations.institution AS institution
, locations.location AS location
, locations.is_unique AS is_unique
FROM locations
WHERE NOT EXISTS (SELECT 1
FROM tissue_data
WHERE tissue_data.locid = locations.locid)
AND NOT EXISTS (SELECT 1
FROM nucacid_data
WHERE nucacid_data.locid = locations.locid);
Table 6.62. Columns in the LOCATIONS_FREE View
| Column | From | Description |
|---|---|---|
| LocId | LOCATIONS.LocId | Identifier for the row |
| Institution | LOCATIONS.Institution | Organization, building, etc. describing the locale of this row's Location |
| Location | LOCATIONS.Location | Specific place/position available for a sample. |
| Is_Unique | LOCATIONS.Is_Unique | Whether or not this location can be used more than once. |
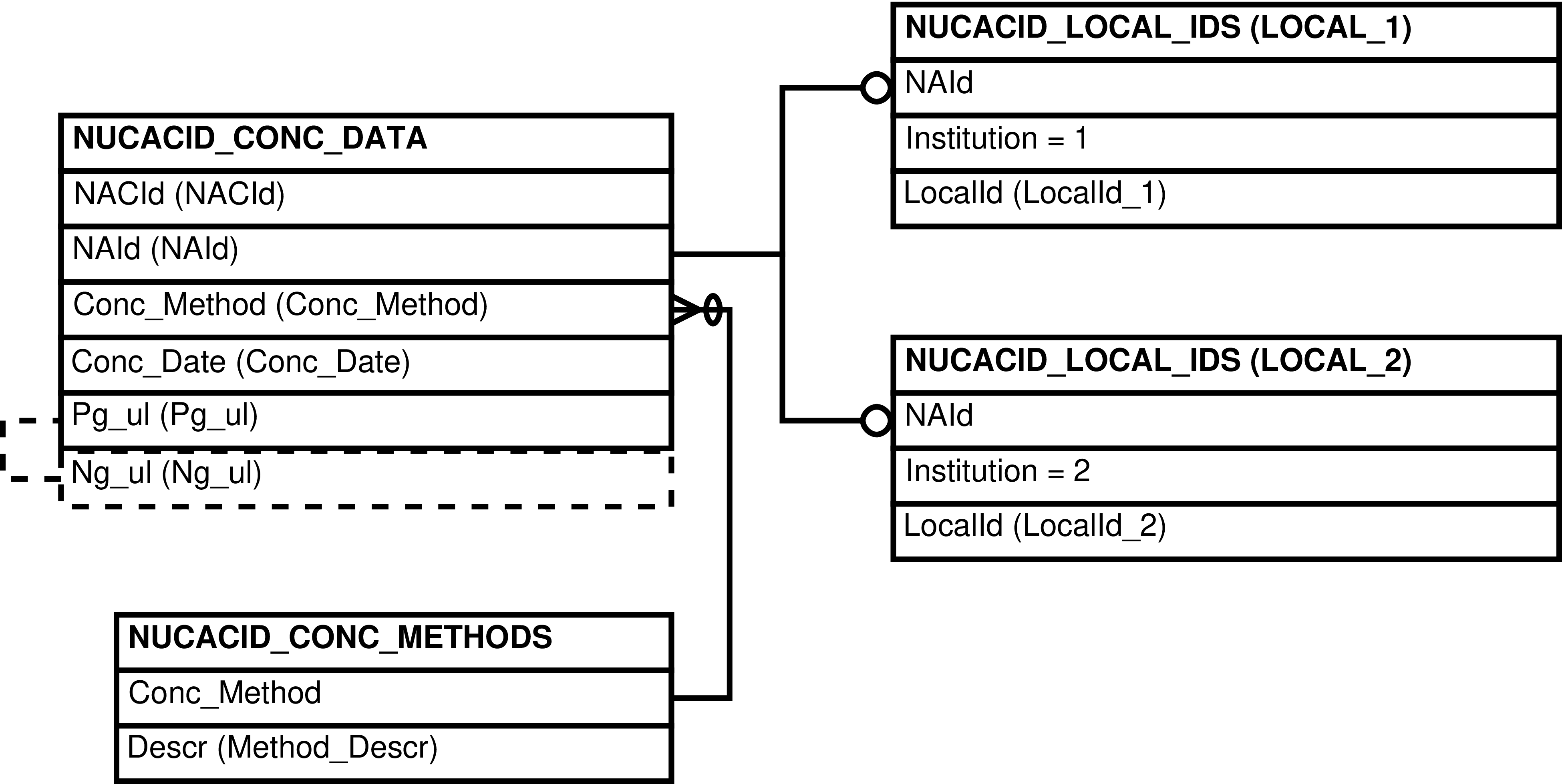
Contains one row for every NUCACID_CONC_DATA row. This view shows all the data from NUCACID_CONC_DATA, but also includes a descriptive column from NUCACID_CONC_METHODS to clarify the meaning of the Conc_Method column.
This view is also useful for adding data. New quantifications can be inserted using a sample's local ID's instead of their NAId.
Tip
Use this view instead of the NUCACID_CONC_DATA table.
Figure 6.132. Query Defining the NUCACID_CONCS View
SELECT nucacid_conc_data.nacid AS nacid
, nucacid_conc_data.naid AS naid
, local_1.localid AS localid_1
, local_2.localid AS localid_2
, nucacid_conc_data.conc_method AS conc_method
, nucacid_conc_methods.descr AS method_descr
, nucacid_conc_data.conc_date AS conc_date
, nucacid_conc_data.quantity AS quantity
, nucacid_conc_data.unit AS unit
FROM nucacid_conc_data
JOIN nucacid_conc_methods
ON nucacid_conc_methods.conc_method = nucacid_conc_data.conc_method
LEFT JOIN nucacid_local_ids AS local_1
ON local_1.naid = nucacid_conc_data.naid
AND local_1.institution = 1
LEFT JOIN nucacid_local_ids AS local_2
ON local_2.naid = nucacid_conc_data.naid
AND local_2.institution = 2;
Table 6.63. Columns in the NUCACID_CONCS View
| Column | From | Description |
|---|---|---|
| NACId | NUCACID_CONC_DATA.NACId | The unique identifier for this quantification. |
| NAId | NUCACID_CONC_DATA.NAId | The unique identifier for the quantified sample. |
| LocalId_1 | NUCACID_LOCAL_IDS.LocalId | The local identifier used for this sample at Institution #1, if any. |
| LocalId_2 | NUCACID_LOCAL_IDS.LocalId | The local identifier used for this sample at Institution #2, if any. |
| Conc_Method | NUCACID_CONC_DATA.Conc_Method | The method of quantification used to determine this concentration. |
| Method_Descr | NUCACID_CONC_METHODS.Descr | A textual description of the quantification method used to determine this concentration. |
| Conc_Date | NUCACID_CONC_DATA.Conc_Date | The date of the quantification. |
| Quantity | NUCACID_CONC_DATA.Quantity | The first half of the concentration: the number. |
| Unit | NUCACID_CONC_DATA.Unit | The second half of the concentration: the unit. |
Inserting a row into NUCACID_CONCS inserts a row into NUCACID_CONC_DATA as expected.
If no NAId is provided, one or both LocalId columns can be provided instead to look up the intended NAId. If LocalId_1 and/or LocalId_2 values are provided, these must be related to a single NUCACID_LOCAL_IDS.NAId value. If a NAId value is also provided, it must equal that single NAId that is related to the provided LocalId column(s).
At least one of either the Conc_Method or Method_Descr columns must be provided to determine the correct value to insert into NUCACID_CONC_DATA.Conc_Method. If Method_Descr is provided, it is used to look up the appropriate Conc_Method value from NUCACID_CONC_METHODS. If both are provided, the provided values must be related in NUCACID_CONC_METHODS.
Updating a row in NUCACID_CONCS updates the underlying NUCACID_CONC_DATA row, as discussed below.
The NAId may be updated in this view via updates to the NAId column only. Updates to the LocalId_1 and LocalId_2 columns result in an error[279].
A row's underlying NUCACID_CONC_DATA.Conc_Method may be updated in this view via updates to the Conc_Method or Method_Descr columns. If more than one of these values is updated, all the newly-updated values must be related, as discussed above.
Updating Conc_Method, Conc_Date, Quantity, or Unit updates the columns in the underlying NUCACID_CONC_DATA row, as expected.
Deleting a row from NUCACID_CONCS deletes a row from NUCACID_CONC_DATA as expected.
Contains one row for every NUCACID_SOURCES row. This view includes several subqueries from NUCACID_LOCAL_IDS, so that each sample and source are shown with their "local" identifiers. This view can be used to upload data.
Tip
Use this view instead of the NUCACID_SOURCES table.
Figure 6.134. Query Defining the NUCACID_SOURCES_EXT View
SELECT nucacid_sources.naid AS naid
, na_1.localid AS na_1
, na_2.localid AS na_2
, nucacid_sources.source_naid AS source_naid
, src_1.localid AS src_1
, src_2.localid AS src_2
, nucacid_sources.relationship
FROM nucacid_sources
LEFT JOIN nucacid_local_ids AS na_1
ON na_1.naid = nucacid_sources.naid
AND na_1.institution = 1
LEFT JOIN nucacid_local_ids AS na_2
ON na_2.naid = nucacid_sources.naid
AND na_2.institution = 2
LEFT JOIN nucacid_local_ids AS src_1
ON src_1.naid = nucacid_sources.source_naid
AND src_1.institution = 1
LEFT JOIN nucacid_local_ids AS src_2
ON src_2.naid = nucacid_sources.source_naid
AND src_2.institution = 2;
Table 6.64. Columns in the NUCACID_SOURCES_EXT View
| Column | From | Description |
|---|---|---|
| NAId | NUCACID_SOURCES.NAId | Identifier for this sample. |
| NA_1 | NUCACID_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #1. |
| NA_2 | NUCACID_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #2. |
| Source_NAId | NUCACID_SOURCES.Source_NAId | Identifier for this source. |
| Src_1 | NUCACID_LOCAL_IDS.LocalId | The local identifier, if any, used for this source at Institution #1. |
| Src_2 | NUCACID_LOCAL_IDS.LocalId | The local identifier, if any, used for this source at Institution #2. |
| Relationship | NUCACID_SOURCES.Relationship | A textual description of how this sample is related to this source. |
Inserting a row into NUCACID_SOURCES_EXT inserts a row in NUCACID_SOURCES, as expected.
To identify the nucleic acid sample, only one of the NAId, NA_1, and NA_2 columns must be provided. If more than one is provided, they must match in NUCACID_LOCAL_IDS.
To identify the source, only one of the Source_NAId, Src_1, and Src_2 columns must be provided. If more than one is provided, they must match in NUCACID_LOCAL_IDS.
Updating a row in NUCACID_SOURCES_EXT updates the underlying row in NUCACID_SOURCES, as expected.
If updating the nucleic acid sample, update only one of the NAId, NA_1, or NA_2 columns. The underlying NUCACID_SOURCES.NAId will be updated as expected.
If updating the source, update only one of the Source_NAId, Src_1, or Src_2 columns. The underlying NUCACID_SOURCES.Source_NAId will be updated as expected.
Deleting a row in NUCACID_SOURCES_EXT deletes the underlying row in NUCACID_SOURCES, as expected.
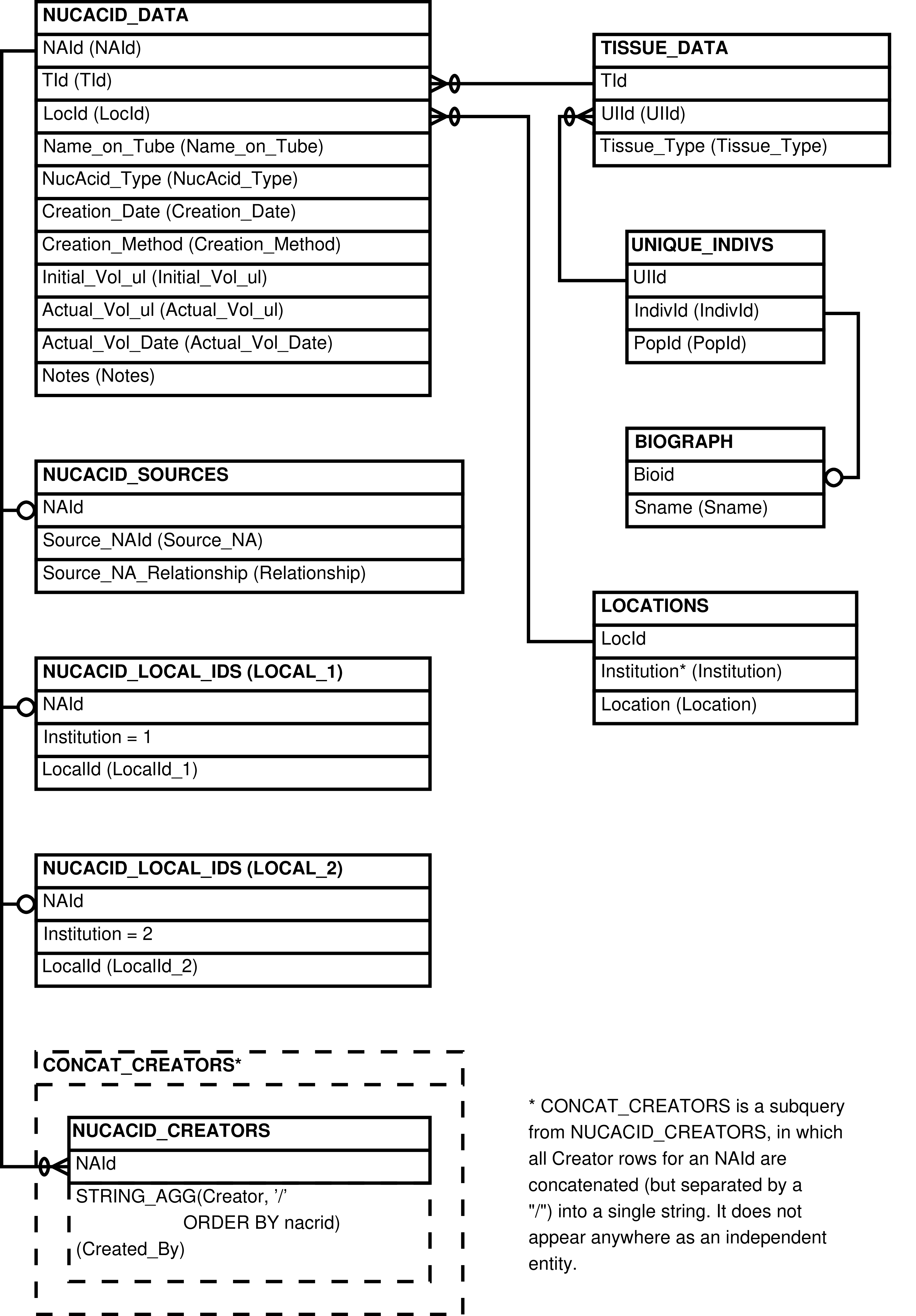
Contains one row for every NUCACID_DATA row. This view includes columns from BIOGRAPH, LOCATIONS, NUCACID_CREATORS, NUCACID_LOCAL_IDS, NUCACID_SOURCES, TISSUE_DATA, and UNIQUE_INDIVS in order to portray information about nucleic acid samples in a more user-friendly format than that in NUCACID_DATA. This view can also be used to upload data.
Tip
Use this view — or the NUCACIDS_W_CONC view if the sample's concentration is important to you — instead of the NUCACID_DATA table.
When uploading data
with this view, it is an error if creator initials cannot be
unambiguously interpreted. In the admittedly-unlikely event
that there is a creator whose initials legitimately include
the separator character
"/", this creator's
initials cannot be inserted via this view. In this case, the
offending creator code must be removed from the data, then
manually inserted into NUCACID_CREATORS.
Figure 6.136. Query Defining the NUCACIDS View
WITH concat_creators AS (SELECT naid
, string_agg(creator, '/' ORDER BY naid, nacrid) AS created_by
FROM nucacid_creators
GROUP BY naid)
SELECT nucacid_data.naid AS naid
, nucacid_data.tid AS tid
, nucacid_data.locid AS locid
, locations.institution AS institution
, locations.location AS location
, local_1.localid AS localid_1
, local_2.localid AS localid_2
, nucacid_data.uiid AS uiid
, unique_indivs.popid AS popid
, unique_indivs.individ AS individ
, biograph.sname AS sname
, nucacid_data.name_on_tube AS name_on_tube
, nucacid_data.nucacid_type AS nucacid_type
, tissue_data.tissue_type AS tissue_type
, nucacid_data.creation_date AS creation_date
, concat_creators.created_by AS created_by
, nucacid_data.creation_method AS creation_method
, COUNT(nucacid_sources.*) AS na_sources
, nucacid_data.initial_vol_ul AS initial_vol_ul
, nucacid_data.actual_vol_ul AS actual_vol_ul
, nucacid_data.actual_vol_date AS actual_vol_date
, nucacid_data.multi_indivs AS multi_indivs
, nucacid_data.multi_tids AS multi_tids
, nucacid_data.notes AS notes
FROM nucacid_data
JOIN locations
ON locations.locid = nucacid_data.locid
LEFT JOIN tissue_data
ON tissue_data.tid = nucacid_data.tid
LEFT JOIN unique_indivs
ON unique_indivs.uiid = nucacid_data.uiid
LEFT JOIN biograph
ON biograph.bioid::text = unique_indivs.individ
AND unique_indivs.popid = 1
LEFT JOIN nucacid_local_ids AS local_1
ON local_1.naid = nucacid_data.naid
AND local_1.institution = 1
LEFT JOIN nucacid_local_ids AS local_2
ON local_2.naid = nucacid_data.naid
AND local_2.institution = 2
LEFT JOIN nucacid_sources
ON nucacid_sources.naid = nucacid_data.naid
LEFT JOIN concat_creators
ON concat_creators.naid = nucacid_data.naid
GROUP BY nucacid_data.naid
, nucacid_data.tid
, nucacid_data.locid
, locations.institution
, locations.location
, local_1.localid
, local_2.localid
, nucacid_data.uiid
, unique_indivs.popid
, unique_indivs.individ
, biograph.sname
, nucacid_data.name_on_tube
, nucacid_data.nucacid_type
, tissue_data.tissue_type
, nucacid_data.creation_date
, concat_creators.created_by
, nucacid_data.creation_method
, nucacid_data.initial_vol_ul
, nucacid_data.actual_vol_ul
, nucacid_data.actual_vol_date
, nucacid_data.multi_indivs
, nucacid_data.multi_tids
, nucacid_data.notes;
Table 6.65. Columns in the NUCACIDS View
| Column | From | Description |
|---|---|---|
| NAId | NUCACID_DATA.NAId | Identifier for this sample. |
| TId | NUCACID_DATA.TId | Identifier for this nucleic acid sample's source tissue sample, if there's only one. |
| LocId | NUCACID_DATA.LocId | Identifier for this sample's Institution-Location pair. |
| Institution | LOCATIONS.Institution | Identifier for this sample's locale. |
| Location | LOCATIONS.Location | The current place/position of the sample. |
| LocalId_1 | NUCACID_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #1. |
| LocalId_2 | NUCACID_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #2. |
| UIId | NUCACID_DATA.UIId | Identifier for the source individual, if recorded. |
| PopId | UNIQUE_INDIVS.PopId | Identifier for the population of the source individual, if the source individual is recorded. |
| IndivId | UNIQUE_INDIVS.IndivId | Name/ID of the source individual, if the source individual is recorded. |
| Sname | BIOGRAPH.Sname | Sname of the source individual, if the source individual is recorded and they have an Sname. |
| Name_on_Tube | NUCACID_DATA.Name_on_Tube | Name/identifier written on the sample's label. |
| NucAcid_Type | NUCACID_DATA.NucAcid_Type | The nucleic acid sample type. |
| Tissue_Type | TISSUE_DATA.Tissue_Type | The source tissue's sample type. |
| Creation_Date | NUCACID_DATA.Creation_Date | Date that the sample was created. |
| Created_By | NUCACID_CREATORS.Creator | Initials of all the personnel involved with
the creation of this sample concatenated into a
single string, ordered by their related NUCACID_CREATORS.NACrId and separated by
a "/". If no
related creators, then NULL. |
| Creation_Method | NUCACID_DATA.Creation_Method | The method used to create the sample. |
| NA_Sources | COUNT(NUCACID_SOURCES.*) | The number of rows in NUCACID_SOURCES with this NAId. |
| Initial_Vol_ul | NUCACID_DATA.Initial_Vol_ul | Volume in microliters of the sample when first created. |
| Actual_Vol_ul | NUCACID_DATA.Actual_Vol_ul | The amount of sample (in microliters) remaining in the tube, as of the Actual_Vol_Date. |
| Actual_Vol_Date | NUCACID_DATA.Actual_Vol_Date | The date that the Actual_Vol_ul was determined. |
| Multi_Indivs | NUCACID_DATA.Multi_Indivs | Boolean indicating whether or not this sample is attributed to more than one individual. |
| Multi_TIds | NUCACID_DATA.Multi_TIds | Boolean indicating whether or not this sample has more than one source tissue. |
| Notes | NUCACID_DATA.Notes | Miscellaneous notes about the sample. |
Inserting a row into NUCACIDS inserts a row into NUCACID_DATA. Additional rows may be inserted into NUCACID_CREATORS or NUCACID_LOCAL_IDS, as discussed below. This view does not insert or update rows in NUCACID_SOURCES.
For each
"/"-separated creator
provided in the Created_By column, one row is inserted
into the NUCACID_CREATORS table, with
the related NAId. A NULL
Created_By column is interpreted to mean that there are no
rows to add to NUCACID_CREATORS; it does
not result in a new NUCACID_CREATORS row with a NULL Creator value.
When either or
both of the LocalId_1 and LocalId_2 columns is not NULL,
a row is inserted into NUCACID_LOCAL_IDS
for each non-NULL value provided. The new NUCACID_LOCAL_IDS.LocalId is the provided
LocalId_N value, and the new Institution is
1 (for LocalId_1) or
2 (for
LocalId_2).
To indicate a sample's current locale and location, either the LocId column or both the Institution and Location columns must be provided. If all three are provided, the Institution and Location must be equal to the related columns in LOCATIONS for the provided LocId.
When the sample should be attributed to a single individual, that individual must be identified via the UIId, PopId, IndivId, and/or Sname columns. It is not necessary to provide values for all of those columns; there must only be enough information provided to identify a single UIId. Specifically: there must be a UIId, a PopId and an IndivId, or an Sname. When more than one of those is provided, all provided values must be related to the same UNIQUE_INDIVS.UIId, which is inserted as the new NUCACIDS.UIId.
When the sample should not be attributed to a single
individual — the NUCACIDS.UIId will be NULL — the
UIId, PopId, IndivId, and Sname must all be NULL.
It is not necessary to provide Tissue_Type. If provided, it must match the related TISSUE_DATA.Tissue_Type value.
The NA_SOURCES column is calculated automatically.
When inserting data, there cannot possibly be any rows in
NUCACID_SOURCES yet, so if a value is
provided for this column it must be
0.
Updating a row in NUCACIDS updates the underlying row in NUCACID_DATA, as expected. Related rows in NUCACID_CREATORS and NUCACID_LOCAL_IDS may be inserted, updated, or deleted, as discussed below. This view does not insert or update rows in NUCACID_SOURCES.
When an update changes the Created_By column, all prior rows in NUCACID_CREATORS are deleted, and new rows are inserted as described above. When an update doesn't change the Created_By column, the related data in NUCACID_CREATORS are unaffected.
When LocalId_1 or LocalId_2 is changed, the related
NUCACID_LOCAL_IDS.LocalId value is also
changed as expected, except when the "old" or "new" value
is NULL. If the change is from NULL to non-NULL, a
new NUCACID_LOCAL_IDS row is inserted,
as discussed above. If
from non-NULL to NULL, the related NUCACID_LOCAL_IDS row is deleted.
Updating the Institution and Location columns updates the related LocId column, as expected.
Updating a sample's UIId can be done by updating the UIId, PopId and IndivId, and/or Sname columns. Any such updates must correspond to exactly one UIId, as discussed above.
The NA_SOURCES column is calculated automatically and cannot be manually changed. When updating data, any value provided for this column is silently ignored.
Deleting a row from NUCACIDS deletes the underlying row from NUCACID_DATA, as expected. Related rows in NUCACID_CREATORS, NUCACID_LOCAL_IDS, and NUCACID_SOURCES, if any, are also deleted.
This view contains one row for every row in NUCACID_DATA. It includes columns from BIOGRAPH, LOCATIONS, NUCACID_CREATORS, NUCACID_LOCAL_IDS, NUCACID_SOURCES, TISSUE_DATA, and UNIQUE_INDIVS, as in the NUCACIDS view. It also includes several additional columns derived from NUCACID_CONC_DATA that indicate the sample's concentration according to various specific quantification methods.
Tip
Use this view — or just NUCACIDS if the sample's concentration is not important to you — instead of the NUCACID_DATA table.
Warning
A nucleic acid sample's concentration may be
quantified more than once with the same method, so
this view shows only the concentration from the most recent
NUCACID_CONC_DATA.Conc_Date for each
method. Because of this, concentrations whose related Conc_Date is NULL are not
included in this view.
Figure 6.138. Query Defining the NUCACIDS_W_CONC View
WITH last_quants AS (SELECT DISTINCT
naid
, conc_method
, last_value(quantity) OVER w AS lastquantity
, last_value(unit) OVER w AS lastunit
, last_value(conc_date) OVER w AS lastdate
FROM nucacid_conc_data
WHERE conc_date IS NOT NULL
WINDOW w AS (PARTITION BY naid, conc_method
ORDER BY conc_date
RANGE BETWEEN UNBOUNDED PRECEDING
AND UNBOUNDED FOLLOWING))
, concat_creators AS (SELECT naid
, string_agg(creator, '/' ORDER BY naid, nacrid) AS created_by
FROM nucacid_creators
GROUP BY naid)
SELECT nucacid_data.naid AS naid
, nucacid_data.tid AS tid
, nucacid_data.locid AS locid
, locations.institution AS institution
, locations.location AS location
, local_1.localid AS localid_1
, local_2.localid AS localid_2
, nucacid_data.uiid AS uiid
, unique_indivs.popid AS popid
, unique_indivs.individ AS individ
, biograph.sname AS sname
, nucacid_data.name_on_tube AS name_on_tube
, nucacid_data.nucacid_type AS nucacid_type
, tissue_data.tissue_type AS tissue_type
, nucacid_data.creation_date AS creation_date
, concat_creators.created_by AS created_by
, nucacid_data.creation_method AS creation_method
, COUNT(nucacid_sources.*) AS na_sources
, nucacid_data.initial_vol_ul AS initial_vol_ul
, nucacid_data.actual_vol_ul AS actual_vol_ul
, nucacid_data.actual_vol_date AS actual_vol_date
, nucacid_data.multi_indivs AS multi_indivs
, nucacid_data.multi_tids AS multi_tids
, nucacid_data.notes AS notes
, CASE
WHEN qpcr.lastquantity IS NOT NULL
THEN convert_conc(qpcr.lastquantity, qpcr.lastunit, 'bb_pgul_unit')
ELSE NULL
END AS qpcr_pg_ul
, qpcr.lastdate AS qpcr_lastdate
, CASE
WHEN nanodrop.lastquantity IS NOT NULL
THEN convert_conc(nanodrop.lastquantity, nanodrop.lastunit, 'bb_ngul_unit')
ELSE NULL
END AS nanodrop_ng_ul
, nanodrop.lastdate AS nanodrop_lastdate
, CASE
WHEN qubit.lastquantity IS NOT NULL
THEN convert_conc(qubit.lastquantity, qubit.lastunit, 'bb_ngul_unit')
ELSE NULL
END AS qubit_ng_ul
, qubit.lastdate AS qubit_lastdate
, CASE
WHEN bioanalyzer.lastquantity IS NOT NULL
THEN convert_conc(bioanalyzer.lastquantity, bioanalyzer.lastunit, 'bb_ngul_unit')
ELSE NULL
END AS bioanalyzer_ng_ul
, bioanalyzer.lastdate AS bioanalyzer_lastdate
, CASE
WHEN quantit.lastquantity IS NOT NULL
THEN convert_conc(quantit.lastquantity, quantit.lastunit, 'bb_ngul_unit')
ELSE NULL
END AS quantit_ng_ul
, quantit.lastdate AS quantit_lastdate
FROM nucacid_data
JOIN locations
ON locations.locid = nucacid_data.locid
LEFT JOIN tissue_data
ON tissue_data.tid = nucacid_data.tid
LEFT JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON biograph.bioid::text = unique_indivs.individ
AND unique_indivs.popid = 1
LEFT JOIN nucacid_local_ids AS local_1
ON local_1.naid = nucacid_data.naid
AND local_1.institution = 1
LEFT JOIN nucacid_local_ids AS local_2
ON local_2.naid = nucacid_data.naid
AND local_2.institution = 2
LEFT JOIN nucacid_sources
ON nucacid_sources.naid = nucacid_data.naid
LEFT JOIN concat_creators
ON concat_creators.naid = nucacid_data.naid
LEFT JOIN last_quants AS qpcr
ON qpcr.conc_method = 1
AND qpcr.naid = nucacid_data.naid
LEFT JOIN last_quants AS nanodrop
ON nanodrop.conc_method = 2
AND nanodrop.naid = nucacid_data.naid
LEFT JOIN last_quants AS qubit
ON qubit.conc_method = 3
AND qubit.naid = nucacid_data.naid
LEFT JOIN last_quants AS bioanalyzer
ON bioanalyzer.conc_method = 4
AND bioanalyzer.naid = nucacid_data.naid
LEFT JOIN last_quants AS quantit
ON quantit.conc_method = 5
AND quantit.naid = nucacid_data.naid
GROUP BY nucacid_data.naid
, nucacid_data.tid
, nucacid_data.locid
, locations.institution
, locations.location
, local_1.localid
, local_2.localid
, nucacid_data.uiid
, unique_indivs.popid
, unique_indivs.individ
, biograph.sname
, nucacid_data.name_on_tube
, nucacid_data.nucacid_type
, tissue_data.tissue_type
, nucacid_data.creation_date
, concat_creators.created_by
, nucacid_data.creation_method
, nucacid_data.initial_vol_ul
, nucacid_data.actual_vol_ul
, nucacid_data.actual_vol_date
, nucacid_data.multi_indivs
, nucacid_data.multi_tids
, nucacid_data.notes
, qpcr.lastquantity
, qpcr.lastunit
, qpcr.lastdate
, nanodrop.lastquantity
, nanodrop.lastunit
, nanodrop.lastdate
, qubit.lastquantity
, qubit.lastunit
, qubit.lastdate
, bioanalyzer.lastquantity
, bioanalyzer.lastunit
, bioanalyzer.lastdate
, quantit.lastquantity
, quantit.lastunit
, quantit.lastdate;
Table 6.66. Columns in the NUCACIDS_W_CONC View
| Column | From | Description |
|---|---|---|
| NAId | NUCACID_DATA.NAId | Identifier for this sample. |
| TId | NUCACID_DATA.TId | Identifier for this nucleic acid sample's source tissue sample, if there's only one. |
| LocId | NUCACID_DATA.LocId | Identifier for this sample's Institution-Location pair. |
| Institution | LOCATIONS.Institution | Identifier for this sample's locale. |
| Location | LOCATIONS.Location | The current place/position of the sample. |
| LocalId_1 | NUCACID_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #1. |
| LocalId_2 | NUCACID_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #2. |
| UIId | NUCACID_DATA.UIId | Identifier for the source individual, if recorded. |
| PopId | UNIQUE_INDIVS.PopId | Identifier for the population of the source individual, if the source individual is recorded. |
| IndivId | UNIQUE_INDIVS.IndivId | Name/ID of the source individual, if the source individual is recorded. |
| Sname | BIOGRAPH.Sname | Sname of the source individual, if the source individual is recorded and they have an Sname. |
| Name_on_Tube | NUCACID_DATA.Name_on_Tube | Name/identifier written on the sample's label. |
| NucAcid_Type | NUCACID_DATA.NucAcid_Type | The nucleic acid sample type. |
| Tissue_Type | TISSUE_DATA.Tissue_Type | The source tissue's sample type. |
| Creation_Date | NUCACID_DATA.Creation_Date | Date that the sample was created. |
| Created_By | NUCACID_CREATORS.Creator | Initials of all the personnel involved with
the creation of this sample concatenated into a
single string, ordered by their related NUCACID_CREATORS.NACrId and separated by
a "/". If no
related creators, then NULL. |
| Creation_Method | NUCACID_DATA.Creation_Method | The method used to create the sample. |
| NA_Sources | COUNT(NUCACID_SOURCES.*) | The number of rows in NUCACID_SOURCES with this NAId. |
| Initial_Vol_ul | NUCACID_DATA.Initial_Vol_ul | Volume in microliters of the sample when first created. |
| Actual_Vol_ul | NUCACID_DATA.Actual_Vol_ul | The amount of sample (in microliters) remaining in the tube, as of the Actual_Vol_Date. |
| Actual_Vol_Date | NUCACID_DATA.Actual_Vol_Date | The date that the Actual_Vol_ul was determined. |
| Multi_Indivs | NUCACID_DATA.Multi_Indivs | Boolean indicating whether or not this sample is attributed to more than one individual. |
| Multi_TIds | NUCACID_DATA.Multi_TIds | Boolean indicating whether or not this sample has more than one source tissue. |
| Notes | NUCACID_DATA.Notes | Miscellaneous notes about the sample. |
| QPCR_Pg_ul | convert_conc(NUCACID_CONC_DATA.Quantity, NUCACID_CONC_DATA.Unit, 'PG/UL') | The concentration of this sample in pg/μL, according to the most recent quantitative PCR. |
| QPCR_LastDate | NUCACID_CONC_DATA.Conc_Date | The date of this row's QPCR_Pg_ul was determined; the date of the most recent QPCR. |
| Nanodrop_Ng_ul | convert_conc(NUCACID_CONC_DATA.Quantity, NUCACID_CONC_DATA.Unit, 'NG/UL') | The concentration of this sample in ng/μL, according to the most recent Nanodrop measurement. |
| Nanodrop_LastDate | NUCACID_CONC_DATA.Conc_Date | The date that this row's Nanodrop_Ng_ul was determined; the date of the most recent Nanodrop measurement. |
| Qubit_Ng_ul | convert_conc(NUCACID_CONC_DATA.Quantity, NUCACID_CONC_DATA.Unit, 'NG/UL') | The concentration of this sample in ng/μL, according to the most recent Qubit measurement. |
| Qubit_LastDate | NUCACID_CONC_DATA.Conc_Date | The date that this row's Qubit_Ng_ul was determined; the date of the most recent Qubit measurement. |
| Bioanalyzer_Ng_ul | convert_conc(NUCACID_CONC_DATA.Quantity, NUCACID_CONC_DATA.Unit, 'NG/UL') | The concentration of this sample in ng/μL, according to the most recent Bioanalyzer run. |
| Bioanalyzer_LastDate | NUCACID_CONC_DATA.Conc_Date | The date that this row's Bioanalyzer_Ng_ul was determined; the date of the most recent Bioanalyzer run. |
| Quantit_Ng_ul | convert_conc(NUCACID_CONC_DATA.Quantity, NUCACID_CONC_DATA.Unit, 'NG/UL') | The concentration of this sample in ng/μ/L, according to the most recent Quant-iT assay. |
| Quantit_LastDate | NUCACID_CONC_DATA.Conc_Date | The date that this row's Quantit_Ng_ul was determined; the date of the most-recent Quant-iT assay. |
Contains one row for every TISSUE_SOURCES row. This view includes several subqueries from TISSUE_LOCAL_IDS, so that each sample and source are shown with their "local" identifiers. This view can be used to upload data.
Tip
Use this view instead of the TISSUE_SOURCES table.
Figure 6.140. Query Defining the TISSUE_SOURCES_EXT View
SELECT tissue_sources.tid AS tid
, t_1.localid AS t_1
, t_2.localid AS t_2
, tissue_sources.source_tid AS source_tid
, src_1.localid AS src_1
, src_2.localid AS src_2
, tissue_sources.relationship
FROM tissue_sources
LEFT JOIN tissue_local_ids AS t_1
ON t_1.tid = tissue_sources.tid
AND t_1.institution = 1
LEFT JOIN tissue_local_ids AS t_2
ON t_2.tid = tissue_sources.tid
AND t_2.institution = 2
LEFT JOIN tissue_local_ids AS src_1
ON src_1.tid = tissue_sources.source_tid
AND src_1.institution = 1
LEFT JOIN tissue_local_ids AS src_2
ON src_2.tid = tissue_sources.source_tid
AND src_2.institution = 2;
Table 6.67. Columns in the TISSUE_SOURCES_EXT View
| Column | From | Description |
|---|---|---|
| TId | TISSUE_SOURCES.TId | Identifier for this sample. |
| T_1 | TISSUE_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #1. |
| T_2 | TISSUE_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #2. |
| Source_TId | TISSUE_SOURCES.Source_TId | Identifier for this source. |
| Src_1 | TISSUE_LOCAL_IDS.LocalId | The local identifier, if any, used for this source at Institution #1. |
| Src_2 | TISSUE_LOCAL_IDS.LocalId | The local identifier, if any, used for this source at Institution #2. |
| Relationship | TISSUE_SOURCES.Relationship | A textual description of how this sample is related to this source. |
Inserting a row into TISSUE_SOURCES_EXT inserts a row in TISSUE_SOURCES, as expected.
To identify the tissue sample, only one of the TId, T_1, and T_2 columns must be provided. If more than one is provided, they must match in TISSUE_LOCAL_IDS.
To identify the source, only one of the Source_TId, Src_1, and Src_2 columns must be provided. If more than one is provided, they must match in TISSUE_LOCAL_IDS.
Updating a row in TISSUE_SOURCES_EXT updates the underlying row in TISSUE_SOURCES, as expected.
If updating the tissue sample, update only one of the TId, T_1, or T_2 columns. The underlying TISSUE_SOURCES.TId will be updated as expected.
If updating the source, update only one of the Source_TId, Src_1, or Src_2 columns. The underlying TISSUE_SOURCES.Source_TId will be updated as expected.
Deleting a row in TISSUE_SOURCES_EXT deletes the underlying row in TISSUE_SOURCES, as expected.
Contains one row for every TISSUE_DATA row. This view includes columns from BIOGRAPH, LOCATIONS, TISSUE_LOCAL_IDS, and UNIQUE_INDIVS in order to portray information about tissue samples in a more user-friendly format than that in TISSUE_DATA. This view can also be used to upload data.
Tip
Use this view instead of the TISSUE_DATA table.
Figure 6.142. Query Defining the TISSUES View
SELECT tissue_data.tid AS tid
, tissue_data.locid AS locid
, locations.institution AS institution
, locations.location AS location
, local_1.localid AS localid_1
, local_2.localid AS localid_2
, tissue_data.uiid AS uiid
, unique_indivs.popid AS popid
, unique_indivs.individ AS individ
, biograph.sname AS sname
, tissue_data.name_on_tube AS name_on_tube
, tissue_data.collection_date AS collection_date
, tissue_data.collection_time AS collection_time
, tissue_data.tissue_type AS tissue_type
, tissue_data.storage_medium AS storage_medium
, tissue_data.misid_status AS misid_status
, tissue_data.collection_date_status AS collection_date_status
, tissue_data.multi_indivs AS multi_indivs
, COUNT(tissue_sources.*) AS tissue_sources
, tissue_data.notes AS notes
FROM tissue_data
JOIN locations
ON locations.locid = tissue_data.locid
LEFT JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON biograph.bioid::text = unique_indivs.individ
AND unique_indivs.popid = 1
LEFT JOIN tissue_local_ids AS local_1
ON local_1.tid = tissue_data.tid
AND local_1.institution = 1
LEFT JOIN tissue_local_ids AS local_2
ON local_2.tid = tissue_data.tid
AND local_2.institution = 2
LEFT JOIN tissue_sources
ON tissue_sources.tid = tissue_data.tid
GROUP BY tissue_data.tid
, tissue_data.locid
, locations.institution
, locations.location
, local_1.localid
, local_2.localid
, tissue_data.uiid
, unique_indivs.popid
, unique_indivs.individ
, biograph.sname
, tissue_data.name_on_tube
, tissue_data.collection_date
, tissue_data.collection_time
, tissue_data.tissue_type
, tissue_data.storage_medium
, tissue_data.misid_status
, tissue_data.collection_date_status
, tissue_data.multi_indivs
, tissue_data.notes;
Table 6.68. Columns in the TISSUES View
| Column | From | Description |
|---|---|---|
| TId | TISSUE_DATA.TId | Identifier for this sample. |
| LocId | TISSUE_DATA.LocId | Identifier for this sample's Institution-Location pair. |
| Institution | LOCATIONS.Institution | Identifier for this sample's locale. |
| Location | LOCATIONS.Location | The current place/position of the sample. |
| LocalId_1 | TISSUE_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #1. |
| LocalId_2 | TISSUE_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #2. |
| UIId | TISSUE_DATA.UIId | Identifier for the source individual, if recorded. |
| PopId | UNIQUE_INDIVS.PopId | Identifier for the population of the source individual, if the source individual is recorded. |
| IndivId | UNIQUE_INDIVS.IndivId | Name/ID of the source individual, if the source individual is recorded. |
| Sname | BIOGRAPH.Sname | Sname of the source individual, if the source individual is recorded and they have an Sname. |
| Name_on_Tube | TISSUE_DATA.Name_on_Tube | Name or ID of the source individual, according to the label on the tube. |
| Collection_Date | TISSUE_DATA.Collection_Date | Date that the sample was collected. |
| Collection_Time | TISSUE_DATA.Collection_Time | Time that the sample was collected. |
| Tissue_Type | TISSUE_DATA.Tissue_Type | The tissue sample type. |
| Storage_Medium | TISSUE_DATA.Storage_Medium | The medium used for storing the sample. |
| Misid_Status | TISSUE_DATA.Misid_Status | The mis-identification status of the sample, if the source individual is recorded. |
| Collection_Date_Status | TISSUE_DATA.Collection_Date_Status | The status of this Collection_Date |
| Multi_Indivs | TISSUE_DATA.Multi_Indivs | Boolean indicating whether this sample has tissue sources from more than one individual. |
| Tissue_Sources | COUNT(TISSUE_SOURCES.*) | The number of rows in TISSUE_SOURCES with this TId. |
| Notes | TISSUE_DATA.Notes | Miscellaneous notes about the sample. |
Inserting a row into TISSUES inserts a row into TISSUE_DATA. Additional rows may be inserted into TISSUE_LOCAL_IDS, as discussed below.
When either or both
of the LocalId_1 and LocalId_2 columns is not NULL, a
row is inserted into TISSUE_LOCAL_IDS
for each non-NULL value provided. The new TISSUE_LOCAL_IDS.LocalId is the provided
LocalId_N value, and the new Institution is
1 (for LocalId_1) or
2 (for
LocalId_2).
It is not necessary to provide all of the UIId, PopId, IndivId, and Sname columns; there must only be enough information provided to identify a single UIId. Specifically: there must be a UIId, a PopId and an IndivId, or an Sname. When more than one of those is provided, all provided values must be related to the same UNIQUE_INDIVS.UIId.
To indicate a sample's current locale and location, either the LocId column or both the Institution and Location columns must be provided. If all three are provided, the Institution and Location must be equal to the related columns in LOCATIONS for the provided LocId.
The Tissue_Sources column is calculated
automatically. When inserting data, there cannot possibly
be any rows in TISSUE_SOURCES yet, so if
a value is provided for this column it must be
0.
Updating a row in TISSUES updates the underlying row in TISSUE_DATA, as expected. Related rows in TISSUE_LOCAL_IDS may be inserted, updated, or deleted, as discussed below.
When LocalId_1 or LocalId_2 is changed, the related
TISSUE_LOCAL_IDS.LocalId value is also
changed, as expected. If this change is from NULL to
non-NULL, a new TISSUE_LOCAL_IDS row
is inserted, as discussed above. If
from non-NULL to NULL, the related TISSUE_LOCAL_IDS row is deleted.
Updating a sample's UIId can be done by updating the UIId, PopId and IndivId, and/or Sname columns. Any such updates must correspond to exactly one UIId, as discussed above.
Updating the Institution and Location columns updates the related LocId column, as expected.
The TISSUE_SOURCES column is calculated automatically and cannot be manually changed. When updating data, any value provided for this column is silently ignored.
Deleting a row from TISSUES deletes the underlying row from TISSUE_DATA, as expected. Related rows in TISSUE_LOCAL_IDS and TISSUE_SOURCES, if any, are also deleted.
Contains one row for every TISSUE_DATA row. This view includes columns from BIOGRAPH, LOCATIONS, TISSUE_LOCAL_IDS, UNIQUE_INDIVS, and HORMONE_SAMPLE_DATA in order to portray information about tissue samples in a more user-friendly format than that in TISSUE_DATA, especially samples that are used in hormone analysis. This view is also useful for uploading new tissue samples that will be used for hormone analysis; it provides a way to upload samples into TISSUE_DATA and HORMONE_SAMPLE_DATA simultaneously.
Figure 6.144. Query Defining the TISSUES_HORMONES View
SELECT tissue_data.tid AS tid
, tissue_data.locid AS locid
, locations.institution AS institution
, locations.location AS location
, local_1.localid AS localid_1
, local_2.localid AS localid_2
, tissue_data.uiid AS uiid
, unique_indivs.popid AS popid
, unique_indivs.individ AS individ
, biograph.sname AS sname
, tissue_data.name_on_tube AS name_on_tube
, tissue_data.collection_date AS collection_date
, tissue_data.collection_time AS collection_time
, tissue_data.tissue_type AS tissue_type
, tissue_data.storage_medium AS storage_medium
, tissue_data.misid_status AS misid_status
, tissue_data.collection_date_status AS collection_date_status
, tissue_data.multi_indivs AS multi_indivs
, COUNT(tissue_sources.*) AS tissue_sources
, tissue_data.notes AS notes
, hormone_sample_data.hsid AS hsid
, hormone_sample_data.fzdried_date AS fzdried_date
, hormone_sample_data.sifted_date AS sifted_date
, hormone_sample_data.avail_mass_g AS avail_mass_g
, hormone_sample_data.avail_date AS avail_date
, hormone_sample_data.comments AS comments
FROM tissue_data
JOIN locations
ON locations.locid = tissue_data.locid
LEFT JOIN unique_indivs
ON unique_indivs.uiid = tissue_data.uiid
LEFT JOIN biograph
ON biograph.bioid::text = unique_indivs.individ
AND unique_indivs.popid = 1
LEFT JOIN tissue_local_ids AS local_1
ON local_1.tid = tissue_data.tid
AND local_1.institution = 1
LEFT JOIN tissue_local_ids AS local_2
ON local_2.tid = tissue_data.tid
AND local_2.institution = 2
LEFT JOIN tissue_sources
ON tissue_sources.tid = tissue_data.tid
LEFT JOIN hormone_sample_data
ON hormone_sample_data.tid = tissue_data.tid
GROUP BY tissue_data.tid
, tissue_data.locid
, locations.institution
, locations.location
, local_1.localid
, local_2.localid
, tissue_data.uiid
, unique_indivs.popid
, unique_indivs.individ
, biograph.sname
, tissue_data.name_on_tube
, tissue_data.collection_date
, tissue_data.collection_time
, tissue_data.tissue_type
, tissue_data.storage_medium
, tissue_data.misid_status
, tissue_data.collection_date_status
, tissue_data.multi_indivs
, tissue_data.notes
, hormone_sample_data.hsid
, hormone_sample_data.fzdried_date
, hormone_sample_data.sifted_date
, hormone_sample_data.avail_mass_g
, hormone_sample_data.avail_date
, hormone_sample_data.comments;
Table 6.69. Columns in the TISSUES_HORMONES View
| Column | From | Description |
|---|---|---|
| TId | TISSUE_DATA.TId | Identifier for this sample. |
| LocId | TISSUE_DATA.LocId | Identifier for this sample's Institution-Location pair. |
| Institution | LOCATIONS.Institution | Identifier for this sample's locale. |
| Location | LOCATIONS.Location | The current place/position of the sample. |
| LocalId_1 | TISSUE_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #1. |
| LocalId_2 | TISSUE_LOCAL_IDS.LocalId | The local identifier, if any, used for this sample at Institution #2. |
| UIId | TISSUE_DATA.UIId | Identifier for the source individual, if recorded. |
| PopId | UNIQUE_INDIVS.PopId | Identifier for the population of the source individual, if the source individual is recorded. |
| IndivId | UNIQUE_INDIVS.IndivId | Name/ID of the source individual, if the source individual is recorded. |
| Sname | BIOGRAPH.Sname | Sname of the source individual, if the source individual is recorded and they have an Sname. |
| Name_on_Tube | TISSUE_DATA.Name_on_Tube | Name or ID of the source individual, according to the label on the tube. |
| Collection_Date | TISSUE_DATA.Collection_Date | Date that the sample was collected. |
| Collection_Time | TISSUE_DATA.Collection_Time | Time that the sample was collected. |
| Tissue_Type | TISSUE_DATA.Tissue_Type | The tissue sample type. |
| Storage_Medium | TISSUE_DATA.Storage_Medium | The medium used for storing the sample. |
| Misid_Status | TISSUE_DATA.Misid_Status | The mis-identification status of the sample, if the source individual is recorded. |
| Collection_Date_Status | TISSUE_DATA.Collection_Date_Status | The status of this Collection_Date |
| Multi_Indivs | TISSUE_DATA.Multi_Indivs | Boolean indicating whether this sample has tissue sources from more than one individual. |
| Tissue_Sources | COUNT(TISSUE_SOURCES.*) | The number of rows in TISSUE_SOURCES with this TId. |
| Notes | TISSUE_DATA.Notes | Miscellaneous notes about the sample. |
| HSId | HORMONE_SAMPLE_DATA.HSId | User-generated identifier for the tissue sample. |
| FzDried_Date | HORMONE_SAMPLE_DATA.FzDried_Date | Date the sample was freeze-dried. |
| Sifted_Date | HORMONE_SAMPLE_DATA.Sifted_Date | Date the freeze-dried sample was sifted. |
| Avail_Mass_g | HORMONE_SAMPLE_DATA.Avail_Mass_g | Amount of sample (in g) remaining in the tube, as of the Avail_Date. |
| Avail_Date | HORMONE_SAMPLE_DATA.Avail_Date | Date that the Avail_Mass_g was determined. |
| Comments | HORMONE_SAMPLE_DATA.Comments | Miscellaneous notes/comments about this sample that are relevant only to hormone analysis. |
Because the primary purpose of this view is facilitate working with TISSUE_DATA and HORMONE_SAMPLE_DATA rows simultaneously, this view only allows operations where there is a practical need to operate on the two tables simultaneously[280]. UPDATE is not allowed on TISSUES_HORMONES; there are no needs satisfied nor utility gained from being able to update both tables at once.
Tip
To update data that appear in this view, use the TISSUES or HORMONE_SAMPLES views.
Inserting a row into TISSUES_HORMONES inserts a row into TISSUE_DATA, then a row into HORMONE_SAMPLE_DATA with the same TId. Additional rows may be inserted into TISSUE_LOCAL_IDS, as discussed above.
It is not necessary to provide all of the UIId, PopId, IndivId, and Sname columns, as discussed above.
To indicate a sample's current locale and location, either the LocId column or both the Institution and Location columns must be provided. If all three are provided, the Institution and Location must be equal to the related columns in LOCATIONS for the provided LocId.
The TISSUE_SOURCES column is calculated automatically.
When inserting data, there cannot possibly be any rows in
TISSUE_SOURCES yet, so if a value is
provided for this column it must be
0.
Deleting a row from TISSUES_HORMONES deletes the underlying row from TISSUE_DATA and from HORMONE_SAMPLE_DATA, as expected. Related rows in TISSUE_LOCAL_IDS and TISSUE_SOURCES, if any, are also deleted.